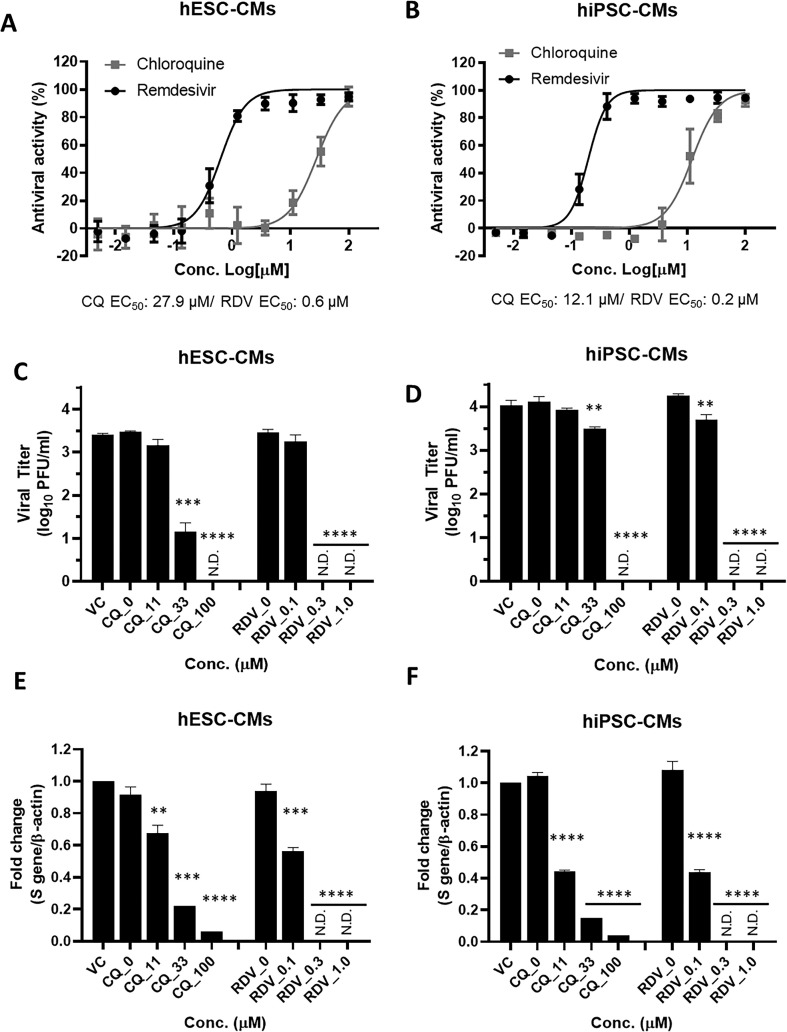
Fig. 4.
Remdesivir potently inhibits SARS-CoV-2 infection in hPSC-CMs. (A and B) Dose-response curve (DRC) analyses of SARS-CoV-2 inhibition by chloroquine and remdesivir in hPSC-CMs. The hESC-CMs (A) and hiPSC-CMs (B) were infected with SARS-CoV-2 at an MOI 2.5 in the presence of various concentrations of chloroquine (grey square) or remdesivir (black circle). At 48 h p.i., the percentage of infected cells was visualized and measured using an automated high-throughput confocal fluorescence imaging system. The data represent the mean (±SD) of at least three independent experiments performed in duplicate. (C to F) Reduction of SARS-CoV-2 replication by chloroquine and remdesivir in hPSC-CMs as determined by infectious viral titer and RT-qPCR. The hESC-CMs (C and E) and hiPSC-CMs (D and F) were infected with SARS-CoV-2 at an MOI of 2.5 in the presence of the indicated concentrations of chloroquine or remdesivir. (C and D) Viral titers from hPSC-CM supernatants were determined with plaque assay in Vero E6 cells. (E and F) Quantification of intracellular SARS-CoV-2 genome RNA by RT-qPCR. Total RNA was isolated from lysates of infected cells for quantification of intracellular SARS-CoV-2 RNA levels (S gene), and results were normalized to β-actin mRNA. Data represent means (±SD) of at least two independent experiments performed in duplicate. Statistical analyses were determined using paired student's t-tests, and significant differences are indicated by **P < 0.01, ***P < 0.005, and ****P < 0.0001. CQ, chloroquine; RDV, remdesivir; N.D, not detected.