Abstract
Background
Studies indicate that the nasal microbiome may correlate strongly with the presence or future risk of childhood asthma.
Objectives
In this study, we tested whether developmental trajectories of the nasopharyngeal microbiome in early life and the composition of the microbiome during illnesses were related to risk of childhood asthma.
Methods
Children participating in the Childhood Origins of Asthma study (N = 285) provided nasopharyngeal mucus samples in the first 2 years of life, during routine healthy study visits (at 2, 4, 6, 9, 12, 18, and 24 months of age), and during episodes of respiratory illnesses, all of which were analyzed for respiratory viruses and bacteria. We identified developmental trajectories of early-life microbiome composition, as well as predominant bacteria during respiratory illnesses, and we correlated these with presence of asthma at 6, 8, 11, 13, and 18 years of age.
Results
Of the 4 microbiome trajectories identified, a Staphylococcus-dominant microbiome in the first 6 months of life was associated with increased risk of recurrent wheezing by age 3 years and asthma that persisted throughout childhood. In addition, this trajectory was associated with the early onset of allergic sensitization. During wheezing illnesses, detection of rhinoviruses and predominance of Moraxella were associated with asthma that persisted throughout later childhood.
Conclusion
In infancy, the developmental composition of the microbiome during healthy periods and the predominant microbes during acute wheezing illnesses are both associated with the subsequent risk of developing persistent childhood asthma.
Key words: Microbiome, children, asthma, development, birth cohort
Abbreviations used: ASV, Amplicon sequence variant; COAST, Childhood Origins of Asthma birth cohort study; GEE, Generalized estimating equation; MPG, Microbiome predominance group; RSV, Respiratory syncytial virus; RV, Rhinovirus
Asthma is a chronic inflammatory disease that affects 6 million children in the United States alone.1 Although childhood asthma can be treated, the lack of a cure underscores the need to understand its early-life developmental origins. Most cases of persistent childhood asthma begin with acute infectious wheezing illnesses in infancy. Although these illnesses are initiated by respiratory viruses, there is strong evidence that bacterial pathogens also contribute,2, 3, 4, 5, 6, 7, 8, 9 and both types of microorganisms have also been related to the subsequent risk of developing asthma. Wheezing illnesses caused by respiratory syncytial virus (RSV) and rhinovirus (RV) are associated with asthma, especially in children who develop early allergic sensitization.10 , 11 In addition, detection of specific bacteria by culture (Streptococcus pneumoniae, Moraxella catarrhalis, or Haemophilus influenzae) or 16S sequencing (eg, Prevotella, Veillonella) in oral or nasopharyngeal aspirates of babies have been related to asthma in early childhood.2 , 12 , 13 In an Australian birth cohort (Childhood Asthma Study) using bacterial metagenomics based on 16S rRNA, predominance of S pneumoniae, M catarrhalis, or H influenzae was found to interact with early allergic sensitization to increase the risk of later asthma.3 , 7 Others have found coassociation between eosinophil counts, severe RV bronchiolitis, and a Haemophilus- or Moraxella-dominated profile of nasopharyngeal microbiota in infants.14 These studies suggest that infection by viral and bacterial pathogens promotes acute wheezing illnesses and increases the risk of asthma while possibly interacting with other host factors such as allergy.
There is also considerable interest in determining whether the dynamic transformation of the airway microbiota with time—its developmental pattern—is associated with acute or chronic respiratory illness. The composition of the airway microbiome typically undergoes marked changes in the first postnatal weeks and months, and this process can be influenced by mode of delivery,15 , 16 viral illnesses17 , 18 and exposure to other children.17 Given the likewise rapid maturation of mucosal immunity in early life, host microbiome dynamics during early childhood may affect future health and disease through interactions with immune development.8 , 19
Collectively, these findings suggest that both the developmental trajectory of the airway microbiome in early life and episodic incursions with viral and bacterial pathogens during respiratory illnesses modify the risk of developing childhood asthma. To further test these hypotheses, we analyzed respiratory bacteria and viruses in nasopharyngeal mucus specimens collected from children enrolled in the Childhood Origins of Asthma (COAST) study under 2 sets of conditions: (1) multiple scheduled visits mostly during periods of good health through 24 months of age and (2) acute respiratory illnesses.20 We derived developmental trajectories of airway microbiome assembly based on the routine samples and then tested these trajectories and microbial composition during respiratory illnesses for associations with asthma throughout childhood.
Methods
Study design
Participants of the COAST birth cohort study (initial N = 289)20 were recruited in Madison, Wisconsin, and surrounding areas from November 1998 to May 2000. The study was approved by the University of Wisconsin-Madison Human Subjects Committee. All families provided informed consent before enrollment, and children provided assent when they reached 7 years of age. All recruited children had at least 1 parent with an allergic disease or asthma. Routine scheduled nasopharyngeal sampling was performed at the time points of 2, 4, 6, 9, 12, 18, and 24 months of age. Most routine samples were collected from children during periods of good health, although some coincided with symptoms of mild respiratory illness. From birth until age 3 years, additional samples were collected from children with upper respiratory illness of at least moderate severity, or with any lower respiratory illness, as previously described.21
The children had yearly routine visits to the clinic where they underwent procedures including assessment of IgE sensitization to aeroallergens (cat, dog, Dermatophagoides pteronyssinus, Dermatophagoide farinae, and Alternaria), blood eosinophil counts, lung function, and asthma diagnosis from ages 6 to 18 years.22 Information on environmental exposures and allergic conditions was collected. Wheezing illnesses, rhinitis, asthma, and atopic dermatitis latent classes were defined as previously described.21 , 23, 24, 25
Detection of viruses and bacteria
We performed 16S rRNA amplicon sequencing of nasopharyngeal samples (swab or aspirate) and negative controls.7 Microbiome data were processed by using QIIME2 (version 2017.10/12)26 and DADA227 to produce relative abundance data for amplicon sequence variants (ASVs), representing unique 16S rRNA V4 sequences. The nasopharyngeal samples were clustered into microbiome profile groups (MPGs) by using hierarchic clustering methods.3 , 7 Nasal specimens were analyzed for common respiratory viruses as previously described.28 , 29
Statistical methods
We used the relative abundances of common ASVs to determine clusters of individuals who shared similar patterns (“trajectories”) of changing microbiome during routine visits (with samples from healthy or mildly ill patients). To generate these trajectories, we omitted all samples obtained at 18 months of age because of the high rate of missing samples at this time point. We then performed multiple-factor analysis (using the R package FactoMineR),30 followed by k-means clustering.
To estimate a longitudinal asthma phenotype, simple latent class models were fit by using asthma diagnoses at ages 6, 8, 11, 13, and 18 years as variables. Next, conditional variable importance measures from random forest ensembles were used to identify microbial and viral features (MPG wheezing burdens, viral wheezing episodes, and routine visit microbiome trajectory) for additional analysis based on associations with the 4-class asthma phenotype.
To compare MPGs and multiple-factor analysis–k-means trajectories, Fisher exact tests and chi-square tests were used for categoric variables; Kruskal-Wallis, t tests, and ANOVA were used for continuous variables. More complex associations were assessed by using generalized linear models for subject-based analyses, or generalized estimating equations (GEEs), using the R package gee [version 4.13.20]31 , 32 for sample-based analyses with adjustment for sex, age, and season with repeated measures of multiple samples per child subject and unstructured correlation. These analyses were conducted by using R software, version 3.5.0. Post hoc comparisons with false discovery rate correction were conducted where required.
Additional details on study and statistical methods are listed in the Online Data Supplement (in this article's Online Repository at www.jacionline.org).
Data and material availability
The microbial sequences have been uploaded to GenBank (accession number pending).
Results
Composition of nasopharyngeal microbiome in COAST
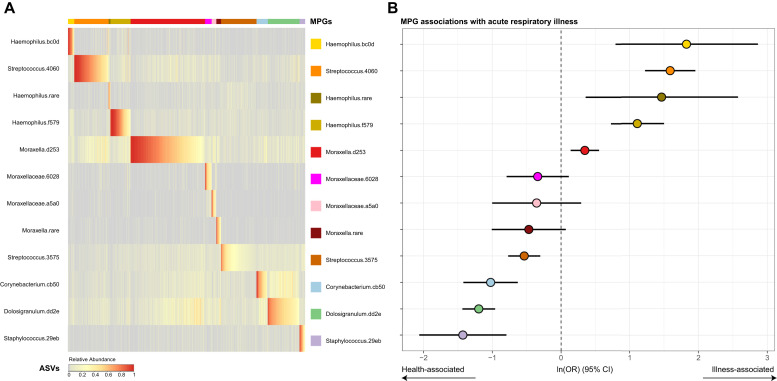
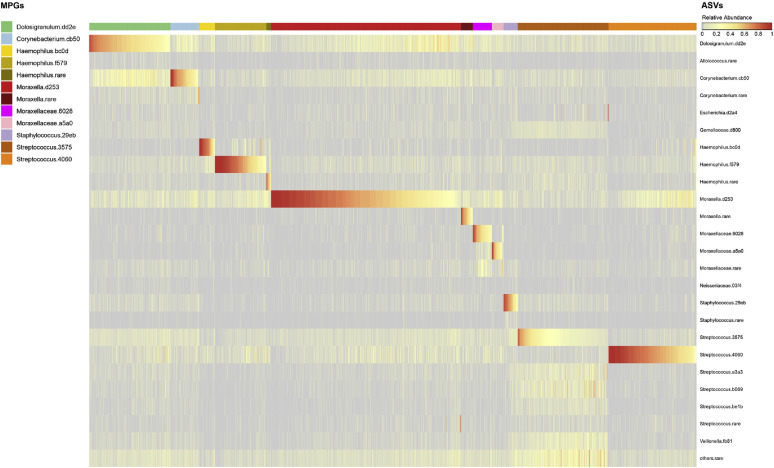
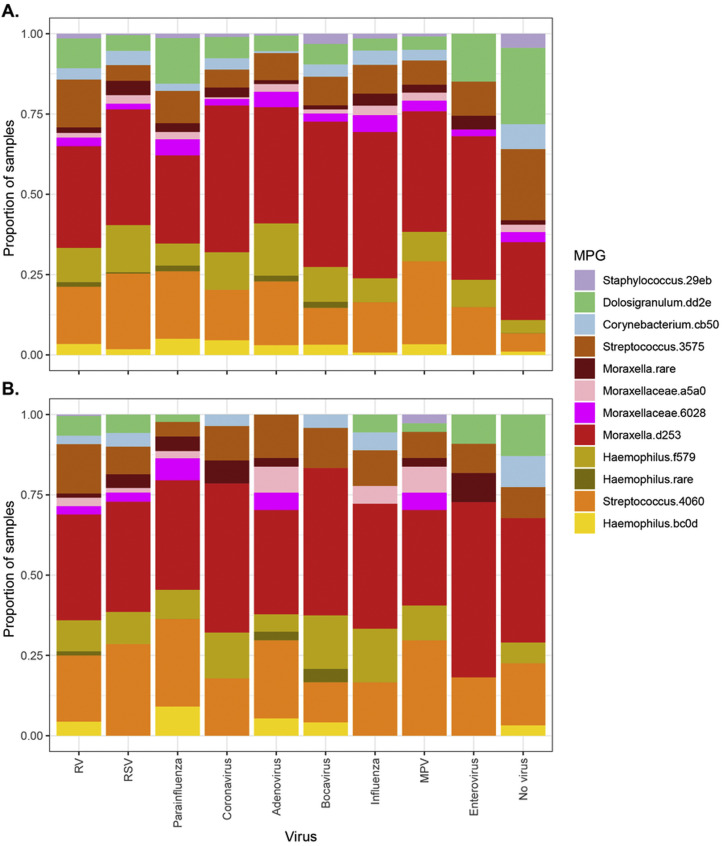
A total of 3147 nasal samples were analyzed for bacteria, including 1654 samples collected during routine scheduled visits (at 2, 4, 6, 9, 12, 18, and 24 months of age) and 1493 additional specimens collected during respiratory illnesses. Of these samples, 2922 passed quality controls (1488 routine and 1434 episodic), of which 1059 were routine and from truly healthy patients, whereas 1863 were samples from patients with illness or mild illness that were collected during either routine or episodic visits. There were 414 distinct samples corresponding to wheezing illnesses. The most common ASVs belonged to 6 genera; Dolosigranulum.dd2e, Corynebacterium.cb50, Haemophilus.bc0d, Haemophilus.f579, Moraxella.d253, Streptococcus.4060, Staphylococcus.29eb, and Streptococcus.3575 (Fig 1 , A). These ASV sequences most closely matched those of the bacterial species Dolosigranulum pigrum, Corynebacterium pseudodiphtheriticum, 2 subtypes of H influenzae, M catarrhalis, S pneumoniae, multiple Staphylococcus species (including S aureus and S epidermidis), and Streptococcus mitis, respectively (see Table E1 in this article's Online Repository at www.jacionline.org).
Fig 1.
Composition of nasopharyngeal microbiome in COAST subjects, and relationship to acute respiratory illness. A, Clustering of microbiomes into microbiome profile groups (MPGs), by relative abundances of ASVs within each sample, as described in the Methods section. The heat in the heatmap represents relative abundance of each ASV (rows, color-coded on the right), arranged by samples (columns) clustered into MPGs (top bar separated by colors of dominant ASV). B, MPG association with respiratory illness, calculated from GEE models with sex, age, and season as covariates. Points (color-coded as per [A]) represent the estimates as natural logarithms of odds ratios (ORs) for association of each MPG with samples from patients with illness versus samples from healthy individualsl, whereas error bars represent 95% CIs for estimates. Numeric results are given in Table E2, A.
Clustering into MPGs
Consistent with previous similar studies,3 , 7 each nasopharyngeal sample had a simple structure, being largely dominated (>50% of reads per sample) by a single ASV. Hierarchic clustering identified 12 MPGs. Each MPG described a pattern with a single dominant ASV and was named according to this dominant taxon. Incidentally, the 12 MPGs corresponded to the most abundant ASVs (Fig 1, A, and for relative abundances for all features, see Fig E1 in this article's Online Repository at www.jacionline.org).
MPG association with acute respiratory illness
Four specific MPGs were significantly overrepresented in the samples from children with respiratory illness compared with in samples from healthy children (P < .05 [Fig 1, B and see Table E2, A in this article's Online Repository at www.jacionline.org). These MPGs were those of known respiratory pathogens Moraxella.d253 (M catarrhalis), Streptococcus.4060 (S pneumoniae), Haemophilus.f579, and Haemophilus.bc0d (both H influenzae). Conversely, MPGs dominated by Corynebacterium.cb50 (C pseudodiphtheriticum), Dolosigranulum.dd2e (D pigrum), Staphylococcus.29eb (Staphylococcus spp), and Streptococcus.3575 (S mitis) were more common in samples from healthy rather than sick children. In a similar analysis testing for association of MPGs with acute wheezing illnesses (compared with samples from healthy children), Streptococcus.4060 (S pneumoniae) showed a significant positive association (P = .00035) and Dolosigranulum.dd23 (D pigrum) showed a negative association (P = 2.6 × 10–5 [see Table E2, A]). Similar results were attained after adjustment for other asthma-related covariates, including parental allergy, parental asthma, environmental smoke exposure, presence of pets, breast-feeding, and birth by Cesarean (see Table E2, B).
As noted in our previous publications,33 , 34 the viruses most commonly detected in the specimens were RV, RSV, parainfluenza virus, coronavirus, and metapneumovirus. We observed that pathogen MPGs (Moraxella.d253, Streptococcus.4060, Haemophilus.f579, and Haemophilus.bc0d) and certain respiratory viruses (RSV and influenza) often coexisted in the same sample, especially during illnesses in the winter months (see Figs E2 and E3 in this article's Online Repository at www.jacionline.org). The distribution of detected MPGs was generally similar across all viruses, whether we examined all samples or only those samples from wheezing illnesses (see Fig E3). During illnesses (n = 1863), pathogen-related MPGs and viruses were most often detected together (n = 1224 [66%]), followed by viruses alone (n = 422 [23%]). pathogen-related MPGs alone (n = 145 [7.8%]), and neither (n = 72 [3.9%]). The presence of any pathogen MPG and the presence of virus each remained independently associated with respiratory illnesses, even with adjustment for each other, age, sex, and seasonality (GEE model, for any pathogen MPG, odds ratio = 3.4 and P = 7.3 × 10–8; for any virus, odds ratio = 12 and P < 1 × 10–10).
Trajectory analysis of the nasopharyngeal microbiome
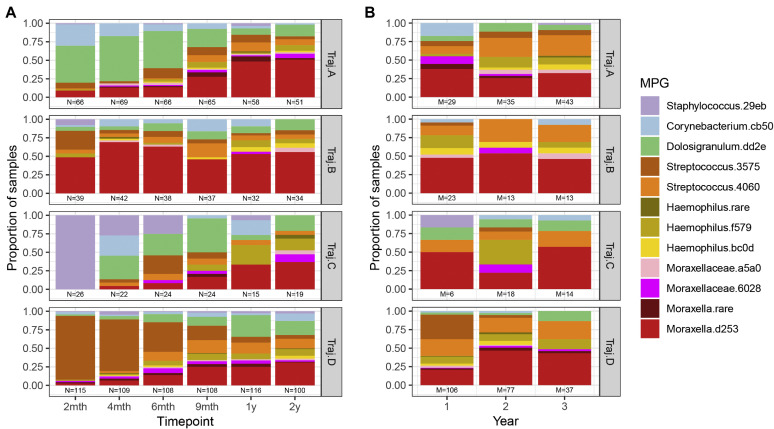
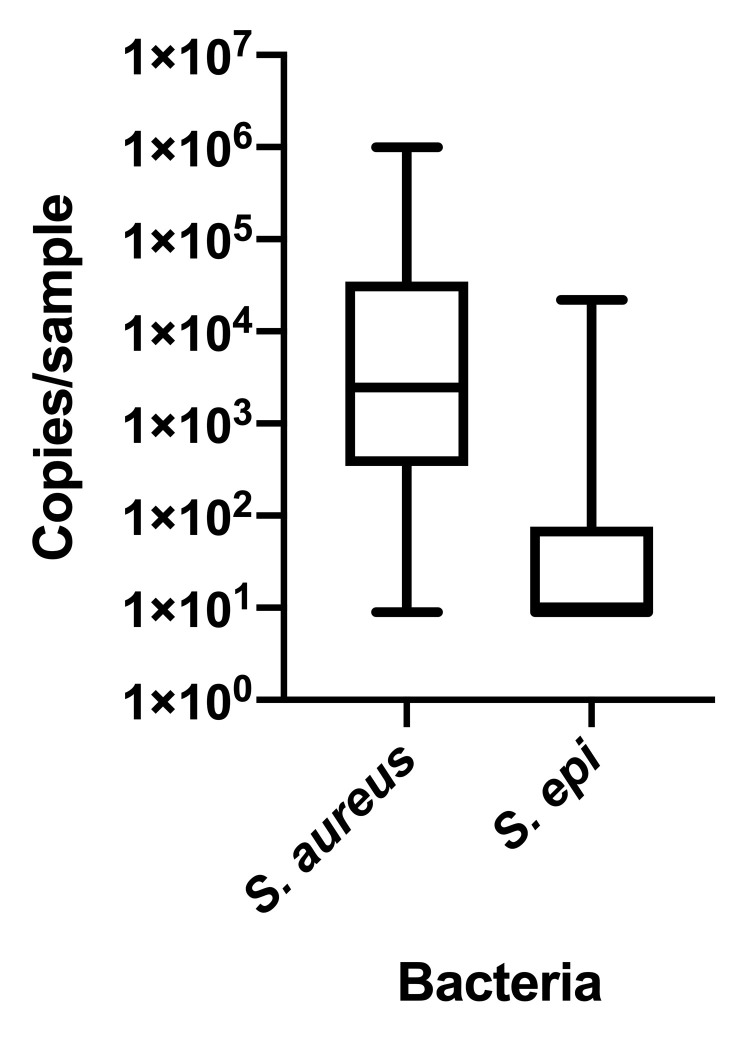
Nasopharyngeal samples from routine study visits across the first 2 years of life were analyzed to identify temporal trajectories of microbiome assembly. We identified 4 clusters of children distinguished by distinct patterns of microbial composition over time (Fig 2 , A). Each trajectory appeared to be driven by a different MPG in the first 6 months of life: trajectory A (n = 79) appeared to be driven by Dolosigranulum.dd2e and Corynebacterium.cb50; trajectory B (n = 43) appeared to be driven by Moraxella.d253; trajectory C (N=26) appeared to be driven by Staphylococcus.29eb; and trajectory D (n = 135) appeared to be driven by Streptococcus.3575 and other streptococci. Because V3 and V4 primers do not reliably differentiate S aureus and S epidermidis, 20 trajectory C nasal mucus samples obtained at 2 months of age were analyzed by quantitative PCR, revealing a predominance of S aureus (see Fig E4 in this article's Online Repository at www.jacionline.org).
Fig 2.
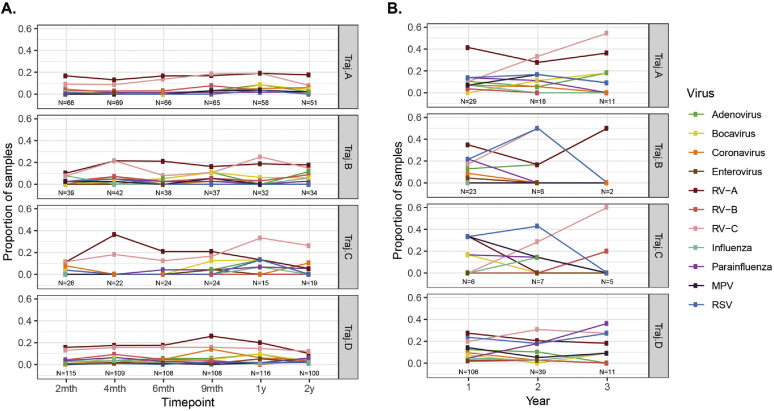
Longitudinal trajectories of nasopharyngeal microbiome. Multiple factor analysis and k-means cluster analysis separated children into trajectories (vertical facets) based on similar patterns of “baseline” microbiome from routine samples from healthy or ill patients in the first 2 years of life. A, Distribution of MPGs as proportions of routine samples (vertical axis) across each trajectory with time point of sampling (horizontal axis). Time points labeled by approximate time of routine visit (eg, 2 months refers to time period spanning 0 to 3 months, 4 months refers to 3 to 5 months, etc). Note the distinctive patterns observed for MPGs in each trajectory, especially in the first 6 months of life. B, Proportion of samples with MPG present during acute wheezing illness in the first 3 years of life among individuals assigned to each baseline, routine sample-based microbiome trajectory as in (A).
Notably, as the children grew older, the trajectories became more similar, and by age 2 years, they had converged toward a generally mixed composition (with many dominated by Moraxella.d253). At age 2 months, between-trajectory dissimilarity (Bray-Curtis) was greatest (0.86), whereas the dissimilarity within the same trajectory was smallest (0.55). These dissimilarities gradually shifted with age, until by age 2 years both between- and within-trajectory dissimilarities were roughly equal (0.71).
During wheezing illnesses, nasal bacteria were typically dominated by illness-associated taxa (eg, Moraxella.d253, Streptococcus.4060, and Haemophilus taxa) irrespective of trajectory (Fig 2, B). There were no significant differences in the rate of detection of specific viruses between any of the 4 trajectories in routine samples or in wheezing illness samples (see Fig E5 in this article's Online Repository at www.jacionline.org).
Demographic characteristics were similar among children in the 4 trajectories (Table I ). There were no significant differences among the microbiome trajectories in terms of other environmental variables, including mode of delivery, presence of pets (cat, dog) in the home, number of siblings at time of birth, exclusive breast-feeding during the first 6 months of life, and antibiotic use in the first year of life (Table I).
Table I.
Demographic characteristics of children in the 4 nasal microbiome trajectories
| Variable | Trajectory |
P value | |||
|---|---|---|---|---|---|
| A |
B |
C |
D |
||
| (n = 79) | (n = 43) | (n = 26) | (n = 135) | ||
| Sex (% male) | 51% | 51% | 69% | 59% | .30 |
| Exclusive breast-feeding for 6 mo | 32% | 30% | 46% | 30% | .43 |
| Dog in home at birth | 42% | 37% | 23% | 34% | .36 |
| Cat at home at birth | 35% | 21% | 35% | 28% | .34 |
| Cesarean delivery | 16% | 12% | 12% | 13% | .91 |
| Maternal asthma ever | 46% | 33% | 35% | 44% | .40 |
| Paternal asthma ever | 36% | 23% | 27% | 29% | .51 |
| Day care in first year | 41% | 53% | 50% | 47% | .54 |
| Nonwhite race | 14% | 9% | 8% | 15% | .75 |
| Mother's education (≥3 years of college) | 73% | 76% | 73% | 70% | .91 |
| Household income ≥ $50,000 | 57% | 68% | 62% | 54% | .43 |
| Older siblings | 52% | 67% | 46% | 55% | .28 |
Association analyses were conducted by using Fisher exact tests for categoric variables, whereas Kruskal tests were conducted for continuous variables across all trajectories.
Association of microbiome trajectories with early wheezing illness and later asthma
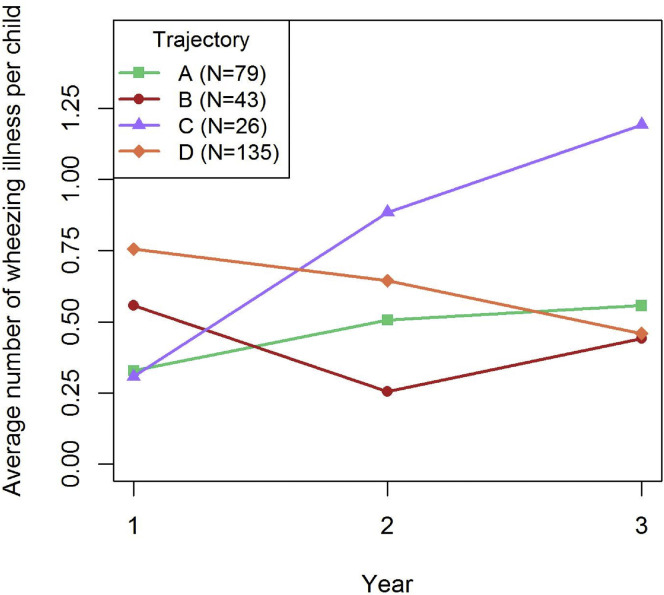
Trajectory C, dominated by Staphylococcus.29eb, was associated with the greatest frequency of wheezing illness in the first 3 years of life; however, this association differed by age (Fig 3 ). The number of wheezing illnesses per trajectory was most similar in the first year of life, lowest for trajectories A (Dolosigranulum) and C (Staphylococcus), and highest for trajectory D (S mitis). However, trajectory C was associated with a progressive increase in wheezing illnesses with time, overtaking the other trajectories to give the greatest frequency at year 3 (P = .0006 for trajectory C).
Fig 3.
Association of nasal microbiome trajectories with the frequency of wheezing illnesses. Number of wheezing illnesses in years 1, 2, and 3 of life was determined for individuals in each of the 4 nasal microbiome trajectories (A, B, C, and D). Microbiome trajectory C, dominated by early Staphylococcus.29eb, was associated with an increase in the number of wheezing illnesses over time (Kruskal test, P = .0006 for trajectory C).
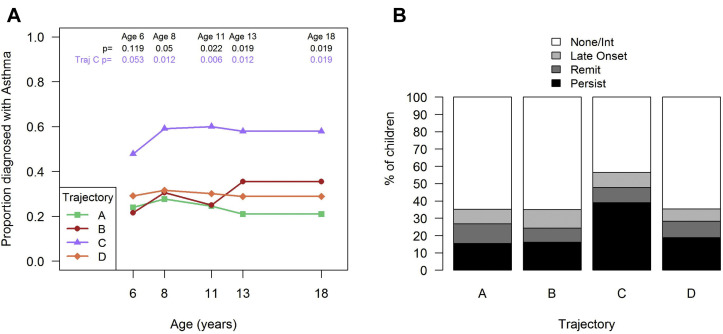
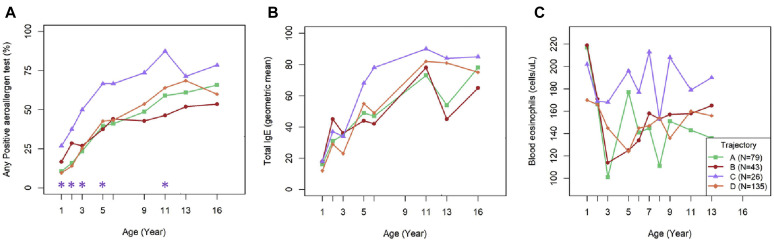
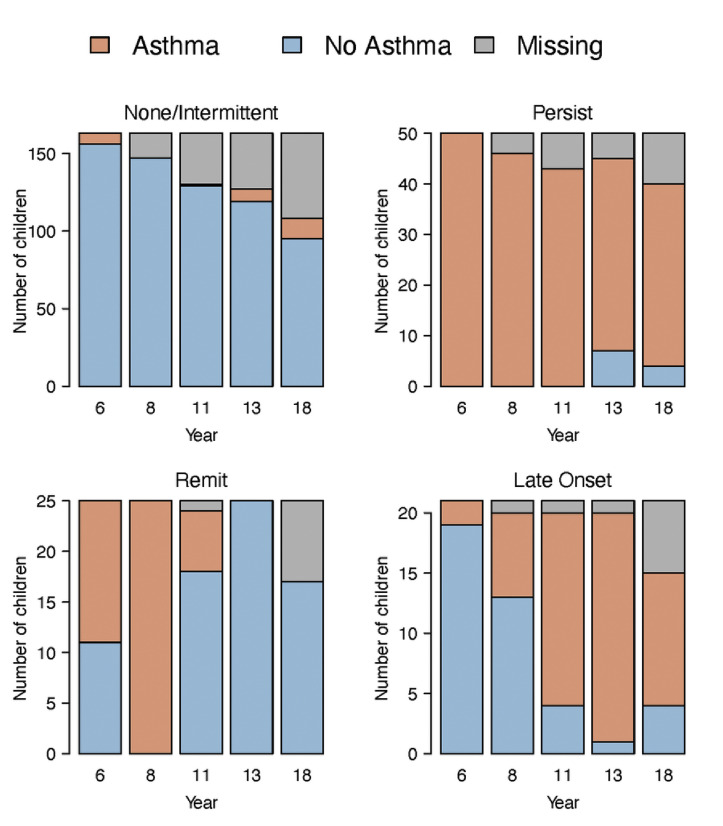
In addition, trajectory C was also associated with greater frequency of physician-diagnosed asthma from age 6 years (47%; P = .053) to 18 years (58%; P = .019) compared with the other trajectories (Fig 4 , A). Furthermore, we applied a latent class model to asthma diagnoses at age 6, 8, 11, 13, and 18 years to identify 4 longitudinal patterns of asthma (see Fig E6 in this article's Online Repository at www.jacionline.org): none or intermittent (63% of subjects), persistent (19% of subjects), remitting (10% of subjects), and late-onset (8% of subjects). Compared with the other microbiome trajectories, trajectory C (Staphylococcus.29eb dominance) tended to be positively associated with a persistent asthma phenotype (P = .08 [Fig 4, B]).
Fig 4.
Association of nasal microbiome trajectories with asthma. Nasal microbiome trajectory C dominated by early Staphylococcus.29eb is associated with higher frequency of asthma at each scheduled assessment (A). P values were obtained by using the chi-square test across all trajectories (top, in black) or post hoc Bonferroni-corrected comparisons for trajectory C versus all other trajectories (A + B + D [bottom, in purple]). Nasal microbiome trajectory C had a higher proportion of children with a persistent asthma phenotype than the other trajectories did (B) (trajectory C vs the other trajectories; P = .08).
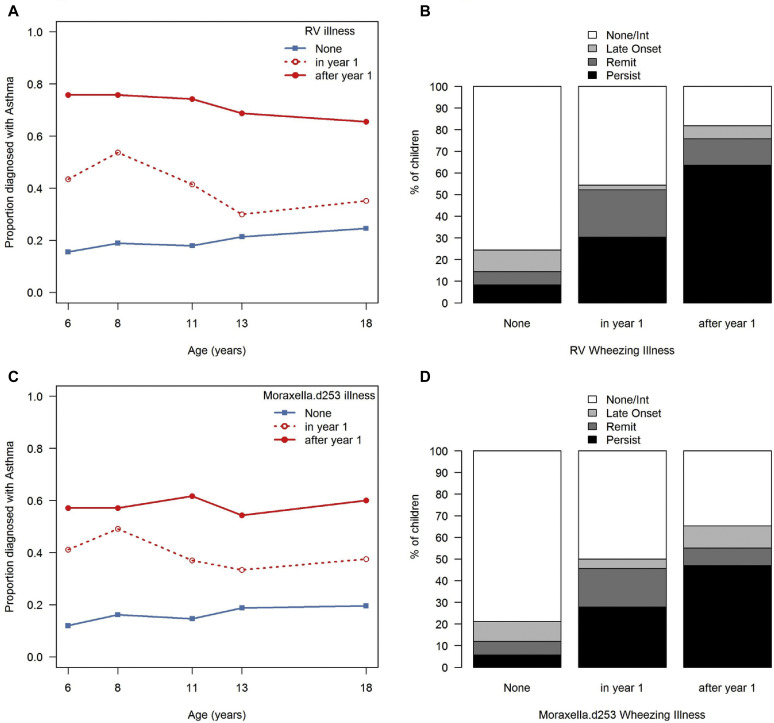
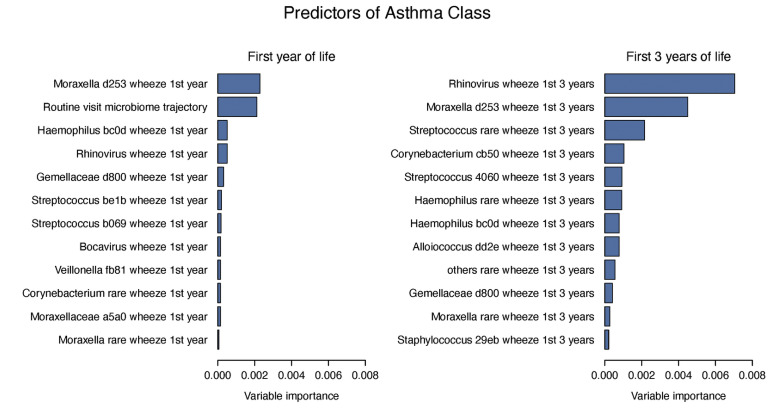
We next evaluated microbial predictors of asthma phenotypes in a random forest model that included the routine visit microbiome trajectories together with MPG and virus detection during wheezing illnesses (see Fig E7 in this article's Online Repository at www.jacionline.org). In the first year of life, microbiome trajectory C along with detection of illness-associated MPGs (Moraxella.d253, Haemophilus.bc0d) were most predictive of asthma class. When the predictors were evaluated over the first 3 years, the microbiome trajectory was no longer among the key predictors of asthma class. Instead, detection of RV during illnesses was an important predictor, and illness-associated MPG Moraxella.d253 remained an important asthma class predictor. These relationships were modified by the age of the child at the time of the wheezing illness (Fig 5 ). Both RV and Moraxella.d253 wheezing illnesses in the first year of life were modestly associated with the persistent asthma latent class, whereas wheezing illnesses associated with RV or Moraxella.d253 during years 2 and 3 were strongly related to persistent asthma.
Fig 5.
Association of microbial pathogen detection during illnesses with asthma. Detection of rhinovirus during wheezing illnesses was associated with increased risks of developing asthma at multiple ages (A). Wheezing illnesses during the second and third years of life were most strongly related to persistent asthma (B). Similar patterns were noted for Moraxella d253 (C and D). P < .001 for all comparisons; Fisher exact test.
Association of microbiome trajectories with allergy variables
Given the close association between early onset of atopy and persistent asthma, we next tested for associations between microbiome trajectory C and indicators of type II inflammation and allergic outcomes. Trajectory C was associated with a greater frequency of aeroallergen sensitization, especially during early childhood (Fig 6 , A). The difference between trajectory C and the others was significant through to age 5 (P < .05 at each age) and also when all years were considered together (trajectory C vs the others; P = .05). There were similar nonsignificant trends for associations between trajectory C and increases in both total IgE level and absolute eosinophil counts (Fig 6, B and C). Trajectory C was associated with a nonsignificant trend for increased risk of allergic rhinitis at age 6 years (overall P = .05, trajectory C vs others P =.12), but not with early-onset atopic dermatitis (see Fig E8 in this article's Online Repository at www.jacionline.org) or lung function (FEV1 or FEV1-to–forced vital capacity ratio [see Table E3 in this article's Online Repository at www.jacionline.org]). A panel of cytokines was analyzed in samples of nasal lavage fluid from a subset of 80 COAST children, with approximately equal representation from each of the 4 MPGs. In general, proinflammatory cytokine production was greatest in the Moraxella MPG, followed by the Staphylococcus, Streptococcus and Dolosigranulum MPGs (see Fig E9 in this article's Online Repository at www.jacionline.org).
Fig 6.
Associations between nasal microbiome trajectories and indicators of atopy. Nasal microbiome trajectory C consistently had a higher proportion of children who were sensitized to at least 1 aeroallergen (A), with similar nonsignificant trends for total IgE (B) and blood eosinophil (C) levels. ∗P < .05 for trajectory C versus other all the trajectories.
We next tested whether the association between trajectory C and asthma was mediated via viral wheezing illnesses or allergic sensitization in early life. To test this, all 3 variables (trajectory, early wheezing illness, and aeroallergen sensitization) were included as outcomes in multivariable models with asthma diagnosis at various time points. The association between trajectory C and asthma diagnoses at ages 6 to 13 was partially ablated with adjustment for both early aeroallergen sensitization (allergen-specific IgE level > 0.35 kU/L by age 2 years) and number of early-life wheezing illnesses up to age 3 (see Table E4 in this article's Online Repository at www.jacionline.org). However, trajectory C remained a statistically significant predictor for asthma diagnosis at ages 11 and 13, suggesting that the microbiome trajectory may be acting via mechanisms not fully captured by wheezing illnesses or early-life aeroallergen sensitization.
The trajectories were robust to modifications in their derivation. We reproduced trajectories by using (1) only routine samples within the first 6 months of life (see the Online Data Supplement) or (2) only healthy samples. Both analyses yielded trajectories that were very similar to the originals (see Table E5 in this article's Online Repository at www.jacionline.org), with similar associations with most asthma outcomes (P < .05 for all generalized linear model associations of asthma at age 8, 11, or 13 years with approximately trajectory C).
Discussion
Developmental patterns of microbiome composition in the gut and skin can influence local immune function and the risk of developing allergic diseases.35, 36, 37 Similarly, we hypothesized that the developmental trajectory of the airway microbiome influences the risk of developing wheezing illnesses and asthma. Children in the COAST study could be separated into 4 developmental trajectories of microbiome composition, each characterized by nasopharyngeal samples in the first 4 to 6 months of life as being dominated by a distinct bacterial taxon. In particular, trajectory C, which was characterized by early Staphylococcus colonization, was associated with a higher frequency of wheezing illnesses during the second and third years of life. Furthermore, membership in the Staphylococcus-dominated trajectory C was linked to increased allergen sensitization, allergic rhinitis, and increased risk of asthma diagnosis from age 6 years through adolescence. The association between trajectory C and asthma was partially mediated by allergic sensitization, RV infections, and early-life wheezing illnesses. Finally, in addition to identifying a novel association between Staphylococcus-dominated nasal microbiome in early life and asthma, we confirmed previously reported relationships between detection of viral (RV)33 and bacterial (eg, M catarrhalis)7 pathogens during periods of illness and the risk of childhood asthma.
Previous observational studies have provided information on temporal changes in composition of the airway microbiome in early life, and both community composition and maturation of the microbiome have been related to more frequent respiratory illnesses. In a study of 60 healthy children sampled several times (at the ages of 1.5, 6, 12, and 24 months) during the first 2 years of age, initial colonization with Haemophilus, Streptococcus, or Staphylococcus communities was associated with more frequent respiratory illnesses and was relatively unstable.16 In contrast, microbial communities associated with Moraxella and Corynebacterium/Dolosigranulum in the first few months were more stable. Our findings were similar in that trajectory B had the most stable composition, with Moraxella MPG detected most often at all ages tested.
The relationship between wheezing illnesses and Staphylococcus appears to be age dependent. Our study and others2 , 7 found that Staphylococcus was more prevalent in secretions from healthy young infants and was less likely to be detected in the first year of life during periods of illness. On the other hand, trajectory C, characterized by the Staphylococcus MPG, was associated with increased wheezing by age 3 years. To reconcile these findings, it is important to consider that the Staphylococcus MPG was predominant in trajectory C only for the first 6 months of life in COAST, and thereafter, Moraxella was the most common MPG. Accordingly, trajectory C was associated with fewer illnesses during the first year, followed by the highest frequency of illnesses during years 2 and 3. Teo et al3 also found that the negative association between Staphylococcus MPG and respiratory illness attenuated over time. Similarly, Bosch et al17 reported that early predominance of Staphylococcus transitioning to Moraxella was related to increased frequency of respiratory illnesses in a birth cohort study. Notably, nasal S aureus has also been related to asthma and bronchial hyperresponsiveness in children38 and wheeze in children and adults.39
There are several potential mechanisms that could link S aureus colonization to childhood asthma. First, S aureus can produce superantigens that are potent stimulators of proinflammatory T-cell responses,40 and can promote type 2 inflammation by directly activating mast cells,41 as well as by inducing thymic stromal lymphopoetin,42 However, analysis of nasal cytokines did not indicate that the S aureus MPG was associated with increased thymic stromal lymphopoetin level or a greater inflammatory milieu in well infants. Alternatively, staphylococci can produce toxins that can enhance viral replication,43 and this effect could lead to increased viral wheezing illnesses. Furthermore, S aureus quorum sensing systems (agr) sense self-produced peptides and upregulate the production of toxins, providing a mechanism for enhanced virulence when S aureus is present in higher quantities.44 , 45 On the skin, S aureus colonization is closely linked to epithelial barrier dysfunction and disease activity in atopic dermatitis and is furthermore associated with a greater risk of sensitization and allergy to foods.46 Accordingly, trajectory C was linked to early aeroallergen sensitization and allergic rhinitis in COAST. Alternatively, considering that Staphylococcus is a predominant organism in the neonatal airway,15 , 47 trajectory C could indicate delayed maturation of the nasal microbiome. Delayed maturation of the microbiome could in turn delay development of airway mucosal immunity and hence lead to more frequent infections.
Detection of pathogen-dominated microbial communities (Moraxella, Streptococcus, Haemophilus, and viruses) have previously been related to acute wheezing illnesses7 , 14 , 17 , 18 , 48 and to childhood asthma at age 5 years.3 , 7 , 49 Similarly, in COAST we found that both RV-associated illnesses and the presence of illness-associated bacteria (especially Moraxella) in nasopharyngeal samples, especially those collected in the second and third years of life, were predictive of persistent childhood asthma. Dumas et al found that a severe bronchiolitis profile characterized by eosinophilia and RV infections is associated with a Moraxella- or Haemophilus-dominated nasopharyngeal microbiota.14 Conversely, Rosas-Salazar et al found that copresence of Lactobacillus during RSV infections may be protective against childhood wheeze.50 These associations suggest that bacterial microbiota during health and disease may influence susceptibility to frequent early-life respiratory infectious illnesses, leading to inflammatory and/or structural changes and entrenchment of asthma. Furthermore, it is possible that there are 2 distinct mechanisms linking the early-life microbiome to asthma: a developmental trajectory that is related to early colonization with Staphylococcus and a second mechanism related to respiratory pathogens (Moraxella, Streptococcus, Haemophilus, and RV) during periods of illness.
Strengths of this study included intensive sampling of the nasal microbiome and virome in the first 2 years of life, which enabled analyses both of microbiome assembly during routinely observed states and during illness-related perturbations. In addition, COAST participants have been evaluated for asthma at regular intervals to the age of 18 years, which enabled identification of children with various patterns of asthma persistence, as well as association of these patterns with microbial traits. One limitation is that COAST participants were specifically selected for family history of asthma and allergy,20 which may limit the generalizability of our findings. In addition, the COAST sample size had limited power to detect associations between environmental factors, microbiome trajectories, and clinical outcomes. The COAST cohort was already assembled before the introduction of conjugated pneumococcal vaccines in 2000, which could have changed patterns of microbial colonization in the upper airways. Finally, the associations in this study link the upper airway microbiome to lower airway outcomes (asthma). Although the upper and lower airway microbiomes have distinct features, close relationships between upper airway microbiome, wheezing, and asthma2, 3, 4, 5, 6, 7 , 51 provide evidence of functional linkages. It is notable that bacteria overexpressed in the lower airways of children and adults with asthma are also commonly present in upper airway samples.52
In summary, these findings suggest that both the initial development of the upper airway microbiome during health and the incursion of specific viral or bacterial pathogens during respiratory illnesses modify the risk of developing persistent childhood asthma. Identifying lifestyle and environmental exposures that promote early colonization with S aureus may lead to future interventional studies to test whether preventing this process can reduce the risk of developing childhood asthma. Another possible opportunity to reduce asthma risk may exist in the form of treatments to prevent infection or proliferation of those major pathogens (eg, RV, M catarrhalis) that are closely associated with acute wheezing illnesses in early life.
Clinical implication.
Identifying factors that promote early colonization with S aureus may lead to future interventional studies to prevent childhood asthma.
Footnotes
Supported by National Institutes of Health, National Heart, Lung, and Blood Institute (grant PO1 HL70381), the National Center for Advancing Translational Sciences (grant UL1TR000427), and the Office of the National Institutes of Health Director (grant no. UG3/UH3 OD023282). M.I. was supported by the Australian National Health and Medical Research Council (grant 1049539). H.H.F.T. was supported by an Australian National Health and Medical Research Council PhD scholarship. K.E.H. was supported by a Senior Medical Research Fellowship from the Viertel Foundation of Victoria.
Disclosure of potential conflict of interest: J. E. Gern is a paid consultant to Ena Therapeutics, Meissa Vaccines, MedImmune, and Regeneron; he has stock options in Meissa Vaccines, as well as a US patent titled “Methods of Propagating Rhinovirus C in Previously Unsusceptible Cell Lines” and a US patent titled “Adapted Rhinovirus C.” The rest of the authors declare that they have no relevant conflicts of interest.
Supplementary data
Fig E1.
Fig E2.
Fig E3.
Fig E4.
Fig E5.
Fig E6.
Fig E7.
Fig E8.
Fig E9.
Fig E10.
References
- 1.Zahran H.S., Bailey C.M., Damon S.A., Garbe P.L., Breysse P.N. Vital signs: asthma in children - United States, 2001-2016. MMWR Morb Mortal Wkly Rep. 2018;67:149–155. doi: 10.15585/mmwr.mm6705e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bisgaard H., Hermansen M.N., Bonnelykke K., Stokholm J., Baty F., Skytt N.L. Association of bacteria and viruses with wheezy episodes in young children: prospective birth cohort study. BMJ. 2010;341:c4978. doi: 10.1136/bmj.c4978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Teo S.M., Tang H.H.F., Mok D., Judd L.M., Watts S.C., Pham K. Airway microbiota dynamics uncover a critical window for interplay of pathogenic bacteria and allergy in childhood respiratory disease. Cell Host Microbe. 2018;24:341–352.e5. doi: 10.1016/j.chom.2018.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kloepfer K.M., Lee W.M., Pappas T.E., Kang T.J., Vrtis R.F., Evans M.D. Detection of pathogenic bacteria during rhinovirus infection is associated with increased respiratory symptoms and asthma exacerbations. J Allergy Clin Immunol. 2014;133:1301–1307.e3. doi: 10.1016/j.jaci.2014.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bashir H., Grindle K., Vrtis R., Vang F., Kang T., Salazar L. Association of rhinovirus species with common cold and asthma symptoms and bacterial pathogens. J Allergy Clin Immunol. 2018;141:822–824.e9. doi: 10.1016/j.jaci.2017.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kloepfer K.M., Sarsani V.K., Poroyko V., Lee W.M., Pappas T.E., Kang T. Community acquired rhinovirus infection is associated with changes in the airway microbiome. J Allergy Clin Immunol. 2017;140:312–315. doi: 10.1016/j.jaci.2017.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Teo S.M., Mok D., Pham K., Kusel M., Serralha M., Troy N. The infant nasopharyngeal microbiome impacts severity of lower respiratory infection and risk of asthma development. Cell Host Microbe. 2015;17:704–715. doi: 10.1016/j.chom.2015.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Steenhuijsen Piters W.A., Heinonen S., Hasrat R., Bunsow E., Smith B., Suarez-Arrabal M.C. Nasopharyngeal microbiota, host transcriptome, and disease severity in children with respiratory syncytial virus infection. Am J Respir Crit Care Med. 2016;194:1104–1115. doi: 10.1164/rccm.201602-0220OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rosas-Salazar C., Shilts M.H., Tovchigrechko A., Schobel S., Chappell J.D., Larkin E.K. Differences in the nasopharyngeal microbiome during acute respiratory tract infection with human rhinovirus and respiratory syncytial virus in infancy. J Infect Dis. 2016;214:1924–1928. doi: 10.1093/infdis/jiw456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kusel M.M., de Klerk N.H., Kebadze T., Vohma V., Holt P.G., Johnston S.L. Early-life respiratory viral infections, atopic sensitization, and risk of subsequent development of persistent asthma. J Allergy Clin Immunol. 2007;119:1105–1110. doi: 10.1016/j.jaci.2006.12.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jackson D.J., Evans M.D., Gangnon R.E., Tisler C.J., Pappas T.E., Lee W.M. Evidence for a causal relationship between allergic sensitization and rhinovirus wheezing in early life. Am J Respir Crit Care Med. 2012;185:281–285. doi: 10.1164/rccm.201104-0660OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thorsen J., Rasmussen M.A., Waage J., Mortensen M., Brejnrod A., Bonnelykke K. Infant airway microbiota and topical immune perturbations in the origins of childhood asthma. Nat Commun. 2019;10:5001. doi: 10.1038/s41467-019-12989-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Powell E.A., Fontanella S., Boakes E., Belgrave D., Shaw A.G., Cornwell E. Temporal association of the development of oropharyngeal microbiota with early life wheeze in a population-based birth cohort. EBioMedicine. 2019;46:486–498. doi: 10.1016/j.ebiom.2019.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dumas O., Hasegawa K., Mansbach J.M., Sullivan A.F., Piedra P.A., Camargo C.A., Jr. Severe bronchiolitis profiles and risk of recurrent wheeze by age 3 years. J Allergy Clin Immunol. 2019;143:1371–1379.e7. doi: 10.1016/j.jaci.2018.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shilts M.H., Rosas-Salazar C., Tovchigrechko A., Larkin E.K., Torralba M., Akopov A. Minimally invasive sampling method identifies differences in taxonomic richness of nasal microbiomes in young infants associated with mode of delivery. Microb Ecol. 2016;71:233–242. doi: 10.1007/s00248-015-0663-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Biesbroek G., Tsivtsivadze E., Sanders E.A., Montijn R., Veenhoven R.H., Keijser B.J. Early respiratory microbiota composition determines bacterial succession patterns and respiratory health in children. Am J Respir Crit Care Med. 2014;190:1283–1292. doi: 10.1164/rccm.201407-1240OC. [DOI] [PubMed] [Google Scholar]
- 17.Bosch A., de Steenhuijsen Piters W.A.A., van Houten M.A., Chu M., Biesbroek G., Kool J. Maturation of the infant respiratory microbiota, environmental drivers, and health consequences. a prospective cohort study. Am J Respir Crit Care Med. 2017;196:1582–1590. doi: 10.1164/rccm.201703-0554OC. [DOI] [PubMed] [Google Scholar]
- 18.Mansbach J.M., Hasegawa K., Henke D.M., Ajami N.J., Petrosino J.F., Shaw C.A. Respiratory syncytial virus and rhinovirus severe bronchiolitis are associated with distinct nasopharyngeal microbiota. J Allergy Clin Immunol. 2016;137:1909–1913.e4. doi: 10.1016/j.jaci.2016.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Folsgaard N.V., Schjorring S., Chawes B.L., Rasmussen M.A., Krogfelt K.A., Brix S. Pathogenic bacteria colonizing the airways in asymptomatic neonates stimulates topical inflammatory mediator release. Am J Respir Crit Care Med. 2013;187:589–595. doi: 10.1164/rccm.201207-1297OC. [DOI] [PubMed] [Google Scholar]
- 20.Lemanske R.F., Jr. The childhood origins of asthma (COAST) study. Pediatr. Allergy Immunol. 2002;13(suppl 15):38–43. doi: 10.1034/j.1399-3038.13.s.15.8.x. [DOI] [PubMed] [Google Scholar]
- 21.Lemanske R.F., Jr., Jackson D.J., Gangnon R.E., Evans M.D., Li Z., Shult P.A. Rhinovirus illnesses during infancy predict subsequent childhood wheezing. J Allergy Clin Immunol. 2005;116:571–577. doi: 10.1016/j.jaci.2005.06.024. [DOI] [PubMed] [Google Scholar]
- 22.Rubner F.J., Jackson D.J., Evans M.D., Gangnon R.E., Tisler C.J., Pappas T.E. Early life rhinovirus wheezing, allergic sensitization, and asthma risk at adolescence. J Allergy Clin Immunol. 2017;139:501–507. doi: 10.1016/j.jaci.2016.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jackson D.J., Gangnon R.E., Evans M.D., Roberg K.A., Anderson E.L., Pappas T.E. Wheezing rhinovirus illnesses in early life predict asthma development in high-risk children. Am J Respir Crit Care Med. 2008;178:667–672. doi: 10.1164/rccm.200802-309OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jackson D.J., Virnig C.M., Gangnon R.E., Evans M.D., Roberg K.A., Anderson E.L. Fractional exhaled nitric oxide measurements are most closely associated with allergic sensitization in school-age children. J Allergy Clin Immunol. 2009;124:949–953. doi: 10.1016/j.jaci.2009.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Singh A.M., Evans M.D., Gangnon R., Roberg K.A., Tisler C., DaSilva D. Expression patterns of atopic eczema and respiratory illnesses in a high-risk birth cohort. J Allergy Clin Immunol. 2010;125:491–493. doi: 10.1016/j.jaci.2009.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.QIIME 2. 2018. Home page. Available at: https://qiime2.org/. Accessed December 18, 2018.
- 27.Callahan B.J., McMurdie P.J., Rosen M.J., Han A.W., Johnson A.J., Holmes S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anderson H.M., Lemanske R.F., Jr., Evans M.D., Gangnon R.E., Pappas T., Grindle K. Assessment of wheezing frequency and viral etiology on childhood and adolescent asthma risk. J Allergy Clin Immunol. 2017;139:692–694. doi: 10.1016/j.jaci.2016.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bochkov Y.A., Grindle K., Vang F., Evans M.D., Gern J.E. Improved molecular typing assay for rhinovirus species A, B, and C. J Clin Microbiol. 2014;52:2461–2471. doi: 10.1128/JCM.00075-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lê S., Josse J., Husson F. FactoMineR: An R package for multivariate analysis. J Stat Soft. 2008;25:18. [Google Scholar]
- 31.Liang K.-Y., Zeger S.L. Longitudinal data analysis using generalized linear models. Biometrika. 1986;73:13–22. [Google Scholar]
- 32.Generalized estimation equation solver. 2015. Available at: https://cran.r-project.org/web/packages/gee/index.html. Accessed December 18, 2018.
- 33.Lee W.M., Lemanske R.F., Jr., Evans M.D., Vang F., Pappas T., Gangnon R. Human rhinovirus species and season of infection determine illness severity. Am J Respir Crit Care Med. 2012;186:886–891. doi: 10.1164/rccm.201202-0330OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gern J.E., Pappas T., Visness C.M., Jaffee K.F., Lemanske R.F., Togias A. Comparison of the etiology of viral respiratory illnesses in inner-city and suburban infants. J Infect Dis. 2012;206:1342–1349. doi: 10.1093/infdis/jis504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fujimura K.E., Sitarik A.R., Havstad S., Lin D.L., Levan S., Fadrosh D. Neonatal gut microbiota associates with childhood multisensitized atopy and T cell differentiation. Nat Med. 2016;22:1187–1191. doi: 10.1038/nm.4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Arrieta M.C., Stiemsma L.T., Dimitriu P.A., Thorson L., Russell S., Yurist-Doutsch S. Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci Transl Med. 2015;7 doi: 10.1126/scitranslmed.aab2271. [DOI] [PubMed] [Google Scholar]
- 37.Kennedy E.A., Connolly J., Hourihane J.O., Fallon P.G., McLean W.H.I., Murray D. Skin microbiome before development of atopic dermatitis: early colonization with commensal staphylococci at 2 months is associated with a lower risk of atopic dermatitis at 1 year. J Allergy Clin Immunol. 2017;139:166–172. doi: 10.1016/j.jaci.2016.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim B.S., Lee E., Lee M.J., Kang M.J., Yoon J., Cho H.J. Different functional genes of upper airway microbiome associated with natural course of childhood asthma. Allergy. 2018;73:644–652. doi: 10.1111/all.13331. [DOI] [PubMed] [Google Scholar]
- 39.Davis M.F., Peng R.D., McCormack M.C., Matsui E.C. Staphylococcus aureus colonization is associated with wheeze and asthma among US children and young adults. J Allergy Clin Immunol. 2015;135:811–813.e5. doi: 10.1016/j.jaci.2014.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Spaulding A.R., Salgado-Pabon W., Kohler P.L., Horswill A.R., Leung D.Y., Schlievert P.M. Staphylococcal and streptococcal superantigen exotoxins. Clin Microbiol Rev. 2013;26:422–447. doi: 10.1128/CMR.00104-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nakamura Y., Oscherwitz J., Cease K.B., Chan S.M., Munoz-Planillo R., Hasegawa M. Staphylococcus delta-toxin induces allergic skin disease by activating mast cells. Nature. 2013;503:397–401. doi: 10.1038/nature12655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vu A.T., Baba T., Chen X., Le T.A., Kinoshita H., Xie Y. Staphylococcus aureus membrane and diacylated lipopeptide induce thymic stromal lymphopoietin in keratinocytes through the Toll-like receptor 2-Toll-like receptor 6 pathway. J Allergy Clin Immunol. 2010;126:985–993.e1-3. doi: 10.1016/j.jaci.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 43.Bin L., Kim B.E., Brauweiler A., Goleva E., Streib J., Ji Y. Staphylococcus aureus alpha-toxin modulates skin host response to viral infection. J Allergy Clin Immunol. 2012;130:683–691.e2. doi: 10.1016/j.jaci.2012.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang R., Braughton K.R., Kretschmer D., Bach T.H., Queck S.Y., Li M. Identification of novel cytolytic peptides as key virulence determinants for community-associated MRSA. Nat Med. 2007;13:1510–1514. doi: 10.1038/nm1656. [DOI] [PubMed] [Google Scholar]
- 45.Baldry M., Nakamura Y., Nakagawa S., Frees D., Matsue H., Nunez G. Application of an agr-specific antivirulence compound as therapy for Staphylococcus aureus-induced inflammatory skin disease. J Infect Dis. 2018;218:1009–1013. doi: 10.1093/infdis/jiy259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jones A.L., Curran-Everett D., Leung D.Y.M. Food allergy is associated with Staphylococcus aureus colonization in children with atopic dermatitis. J Allergy Clin Immunol. 2016;137:1247–1248.e3. doi: 10.1016/j.jaci.2016.01.010. [DOI] [PubMed] [Google Scholar]
- 47.Pattaroni C., Watzenboeck M.L., Schneidegger S., Kieser S., Wong N.C., Bernasconi E. Early-life formation of the microbial and immunological environment of the human airways. cell host microbe. cell Host Microbe. 2018;24:857–865.e4. doi: 10.1016/j.chom.2018.10.019. [DOI] [PubMed] [Google Scholar]
- 48.Hasegawa K., Mansbach J.M., Bochkov Y.A., Gern J.E., Piedra P.A., Bauer C.S. Association of rhinovirus C bronchiolitis and immunoglobulin E sensitization during infancy with development of recurrent wheeze. JAMA Pediatr. 2019;173:544–552. doi: 10.1001/jamapediatrics.2019.0384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bisgaard H., Hermansen M.N., Buchvald F., Loland L., Halkjaer L.B., Bonnelykke K. Childhood asthma after bacterial colonization of the airway in neonates. N Engl J Med. 2007;357:1487–1495. doi: 10.1056/NEJMoa052632. [DOI] [PubMed] [Google Scholar]
- 50.Rosas-Salazar C., Shilts M.H., Tovchigrechko A., Schobel S., Chappell J.D., Larkin E.K. Nasopharyngeal Lactobacillus is associated with a reduced risk of childhood wheezing illnesses following acute respiratory syncytial virus infection in infancy. J Allergy Clin Immunol. 2018;142:1447–1456.e9. doi: 10.1016/j.jaci.2017.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Altman M.C., Gill M.A., Whalen E., Babineau D.C., Shao B., Liu A.H. Transcriptome networks identify mechanisms of viral and nonviral asthma exacerbations in children. Nat Immunol. 2019;20:637–651. doi: 10.1038/s41590-019-0347-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huang Y.J., Nariya S., Lynch S.V., Harris J., Choy D., Arron J.R. The airway microbiome in severe asthma. Ann Am Thorac Soc. 2014;11(suppl 1):S78. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.