Fig. 2. Dimerization of the PRC2 core complex.
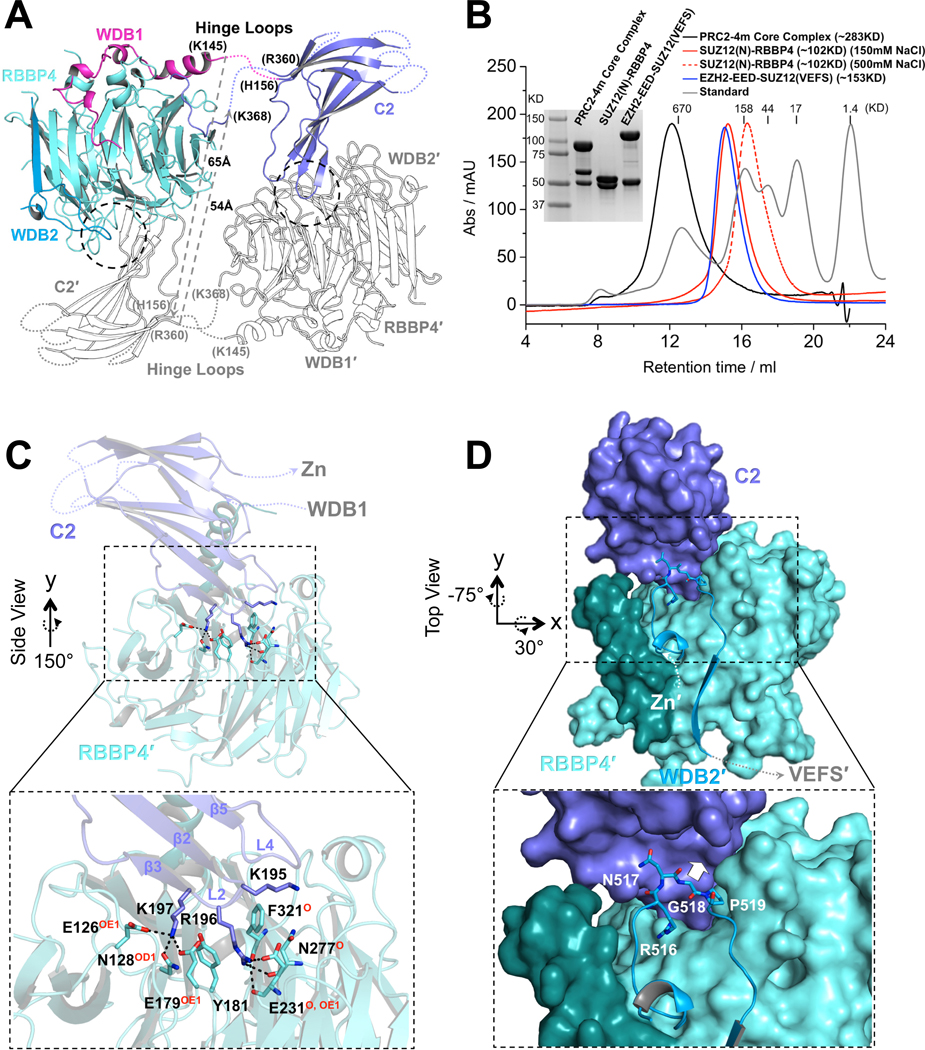
(A) Structure of one protomer of the PRC2 core complex in cartoon representation. Only SUZ12(N) and RBBP4 are shown. Domains within only one protomer are colored. The SUZ12(ZnB) and SUZ12(Zn) domains are removed for clarity. SUZ12 residues at the junction of the disordered hinge loops are labeled. The minimal lengths of a pair of hypothetical hinge loops between the WDB1 domain from one protomer and the C2 domain from the other protomer that would exist in a closed complex are indicated by dotted gray lines.
(B) SEC elution profiles of the four-member PRC2 core complex (PRC2–4m), SUZ12(N)-RBBP4 (at both 150mM and 500mM salt), and EZH2-EED-SUZ12(VEFS). SDS-PAGE gel image is provided. In the last complex, EZH2 and SUZ12(VEFS) were expressed as a fusion protein.
(C) Zoom-in view of the dimer interface between the SUZ12(C2) domain and RBBP4. SUZ12(C2) residues on the dimer interface are shown as sticks. RBBP4 residues involved in SUZ12(C2) binding are indicated, with hydrogen bonding atoms highlighted in red.
(D) Zoom in view of the dimer interface between the SUZ12(C2) and SUZ12(WDB2) domains. Steric clash to the SUZ12(C2) would be imposed by bulky amino acids at residue G518 and is indicated by a white arrow.
See also Fig. S2.
