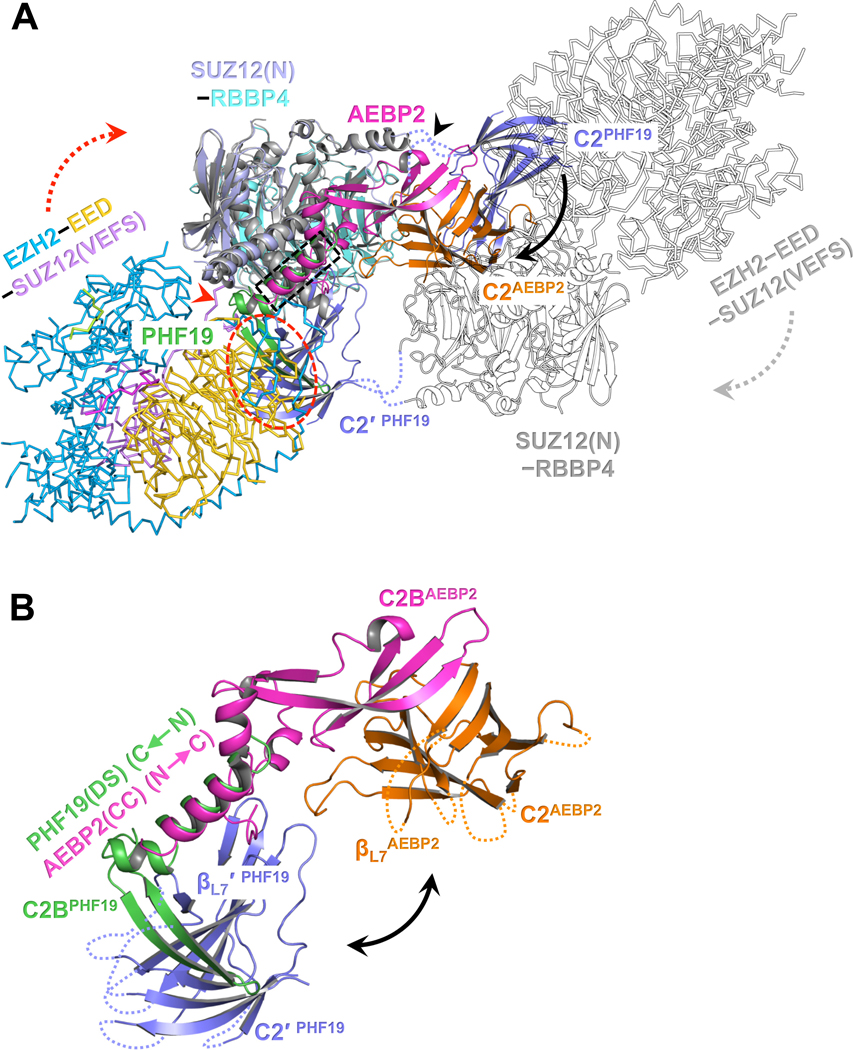
Fig. 4. Distinct structural architectures of the PRC2-PCL and PRC2-AEBP2 complexes.
(A) Structure of PRC2-AEBP2 was constructed by fitting the crystal structures of EZH2-EED-SUZ12(VEFS) (PDB: 5HYN) and SUZ12(N)-RBBP4-AEBP2(C2B-H3K4D) (PDB: 5WAI) into a cryo-EM density map of the corresponding holo complex (EMD-7334). To model the PRC2-PHF19 dimer, PRC2-AEBP2 was structurally aligned to the dimeric SUZ12(N)-RBBP4-PHF19(RC). The two C2 domains in PRC2-PHF19 are colored in purple and labeled as C2PHF19 and C2′ PHF19 (other parts of this protomer are shown as outlines for clarity). The C2 domain PRC2-AEBP2, C2AEBP2, is colored in orange, the AEBP2(C2B-H3K4D) in magenta, and the PHF19(RC) in green.
The black arrow pointing from C2PHF19 to C2AEBP2 indicates the movement of the C2 domain induced by AEBP2 binding that disrupts the intrinsic PRC2 dimer, along the hinge loops indicated by a black arrowhead. The dotted red oval indicates that the C2 domain in PRC2-PHF19 would clash with the EZH2-EED-SUZ12(VEFS) moiety from PRC2-AEBP2. To avoid the steric clash, the EZH2-EED-SUZ12(VEFS) moiety may move along the linker (red arrowhead) between the SUZ12(N) and SUZ12(VEFS) as indicated by the dotted red arrow (and the dotted gray arrow in the other protomer). The dotted black rectangle indicates the overlapped binding surface for the PHF19(DS) and AEBP2(CC) helices.
(B) Zoom-in view of the structural alignment of the SUZ12(C2)-PHF19(RC) and SUZ12(C2)-AEBP2(C2B-H3K4D) interactions. The polarities of the PHF19(DS) and AEBP2(CC) helices are opposite. The black double-headed arrow indicates that the position of the C2 domain of SUZ12 is completely different in PRC2-PHF19 and PRC2-AEBP2.
See also Fig. S4.