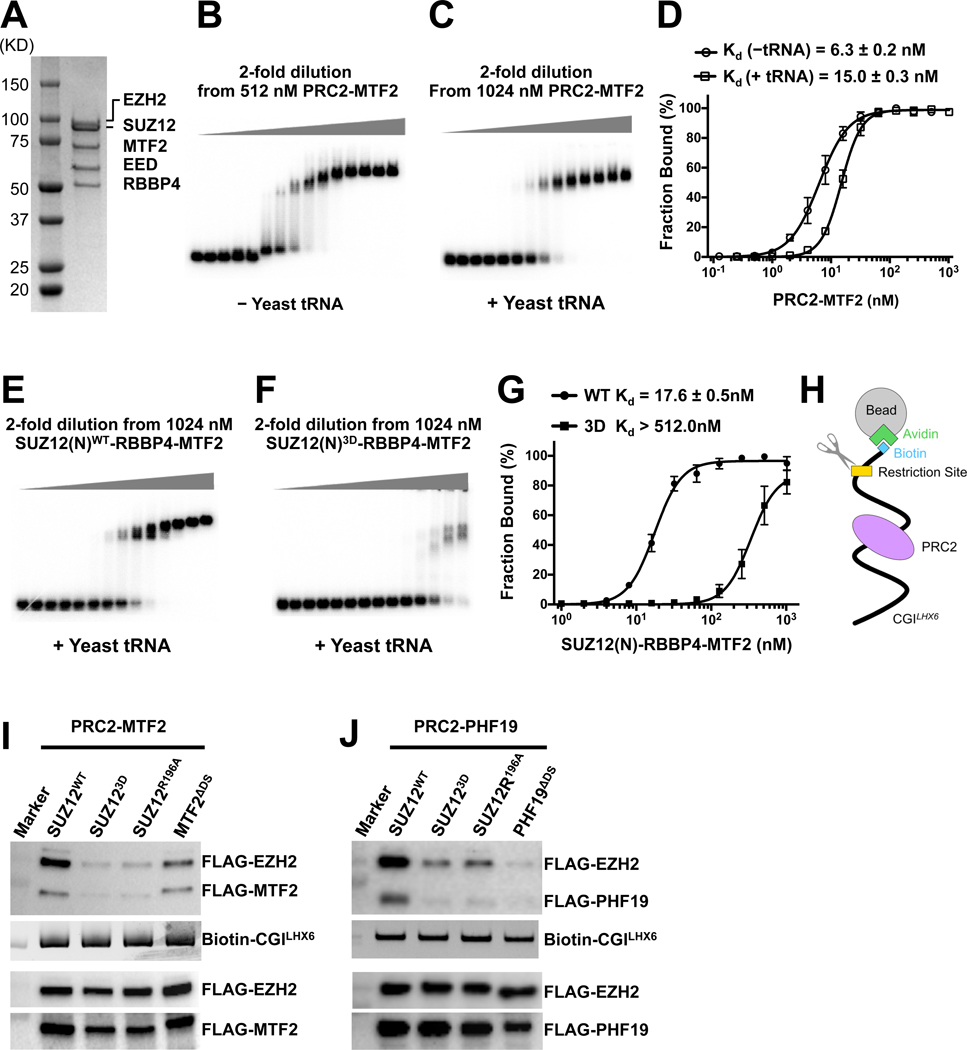
Fig. 5. Determining role of PRC2 dimerization in DNA binding by PRC2-PCL.
(A) SDS-PAGE gel of the reconstituted PRC2-MTF2 holo complex
(B) and (C) EMSA of the binding of the PRC2-MTF2 holo complex to the 32P-labeled CGILHX6 DNA probe in the absence and presence of yeast tRNA.
(D) Quantification of the binding affinities measured in (B) and (C). (D) and (G) were based on three independently performed EMSAs. Graphs display mean ± SEM. GraphPad Prism 8.0 was used for data analysis.
(E) and (F) EMSA of the binding of the SUZ12(N)-RBBP4-MTF2 ternary complex to the CGILHX6 DNA probe in the presence of yeast tRNA. The binding condition is identical to (C). SUZ12WT and SUZ123D were assayed.
(G) Quantification of the binding affinities measured in (E) and (F).
(H) Schematic of the biotinylated DNA pull-down assay.
(I) and (J) Role of PRC2 dimerization in DNA binding. EZH2, EED, MTF2 and PHF19 all contained an N-terminal FLAG tag. Input controls were indicated by anti-FLAG signals (holo complexes, the lower two panels) and SYBR Gold signals (DNA probe, the middle panel). PRC2-MTF2 or PRC2-PHF19 released by the restriction enzyme was detected as anti-FLAG signals (the top panel).
See also Fig. S5.