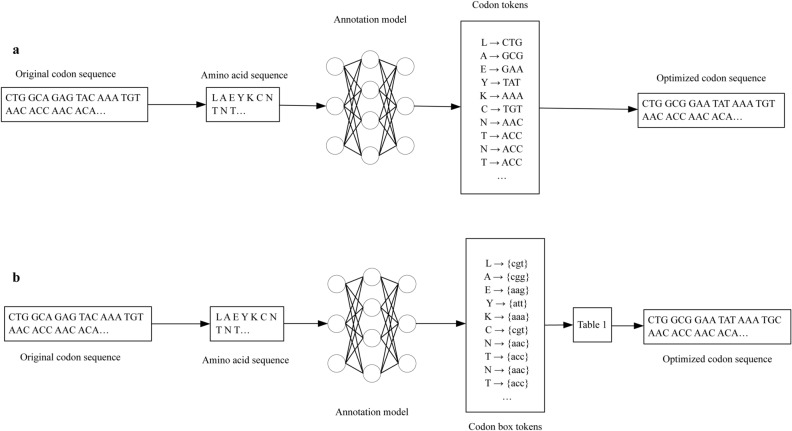
Figure 2.
Codon optimization flowcharts based on sequence annotation models. First, the original codon sequences are decoded into amino acid sequences. Then, they are annotated by the trained sequence annotation models. In the flowchart in (a), the amino acid sequence is annotated with 61 kinds of codons, except stop codons (named BiLSTM-CRF(a)), and in the flowchart in (b), the amino acid sequence is annotated with 20 kinds of codon boxes (named BiLSTM-CRF(b)). The difference in (b) is that the optimized codons are determined from the codon boxes in Table 1 due to the one-to-one mapping of amino acids and codon boxes with codons mentioned in the previous section. Generally, the annotation model with fewer tokens is better, and the complexity of BiLSTM-CRF(b) is lower than that of BiLSTM-CRF(a).