Figure 5.
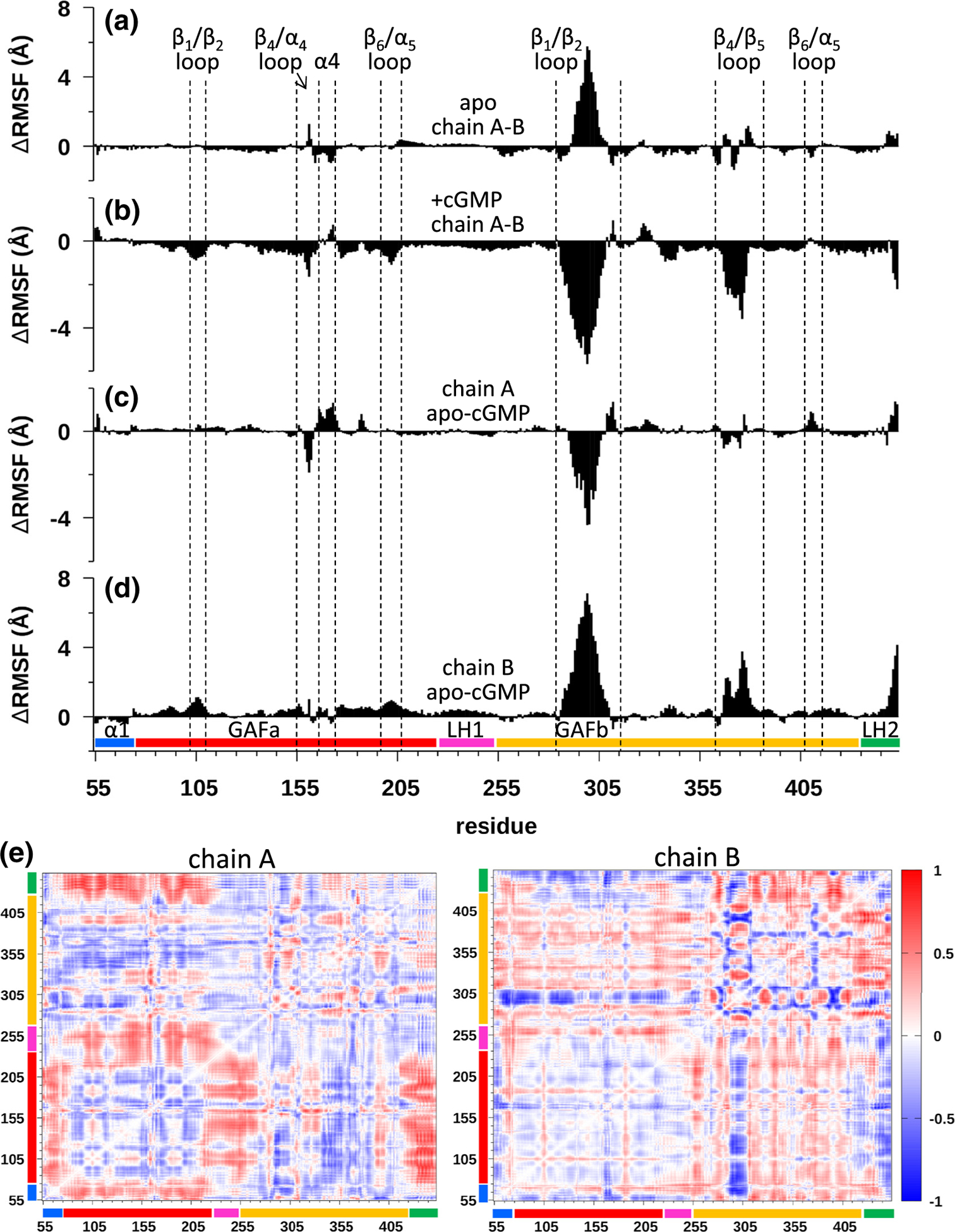
Molecular dynamics (MD) simulations of the apo and cGMP-liganded states of cone PDE6 GAFab. (a)–(d) The ΔRMSF per residue were analyzed separately for each sub-domain of GAFab (N-terminal region (α1) preceding GAFa (residues 55–74), GAFa (residues 75–224), long helix-1 (LH1, residues 225–255), GAFb (residues 256–443), and long helix-2 (LH2, residues 434–453)) to identify differences in protein dynamics of the two chains in the apo and cGMP-bound states for each subunit. (a and b) Evaluation of asymmetry in protein dynamics of the two GAFab subunits in the apo (a) and cGMP-bound (b) state. (c and d) Changes in protein dynamics upon cGMP binding to GAFab were evaluated by plotting the ΔRMSF per residue for the apo and cGMP-liganded states for subunit A (a) and subunit B (d). (e) Difference dynamic cross-correlation (DDCC) analysis of the differences in correlated motions between the apo and cGMP-liganded states of GAFab for subunits A (left) and B (right). Heat map bar indicates the range of correlations from −1 (highly decreased correlation) to 0 (no change in correlation) to +1 (highly increased correlation).
