Figure 1:
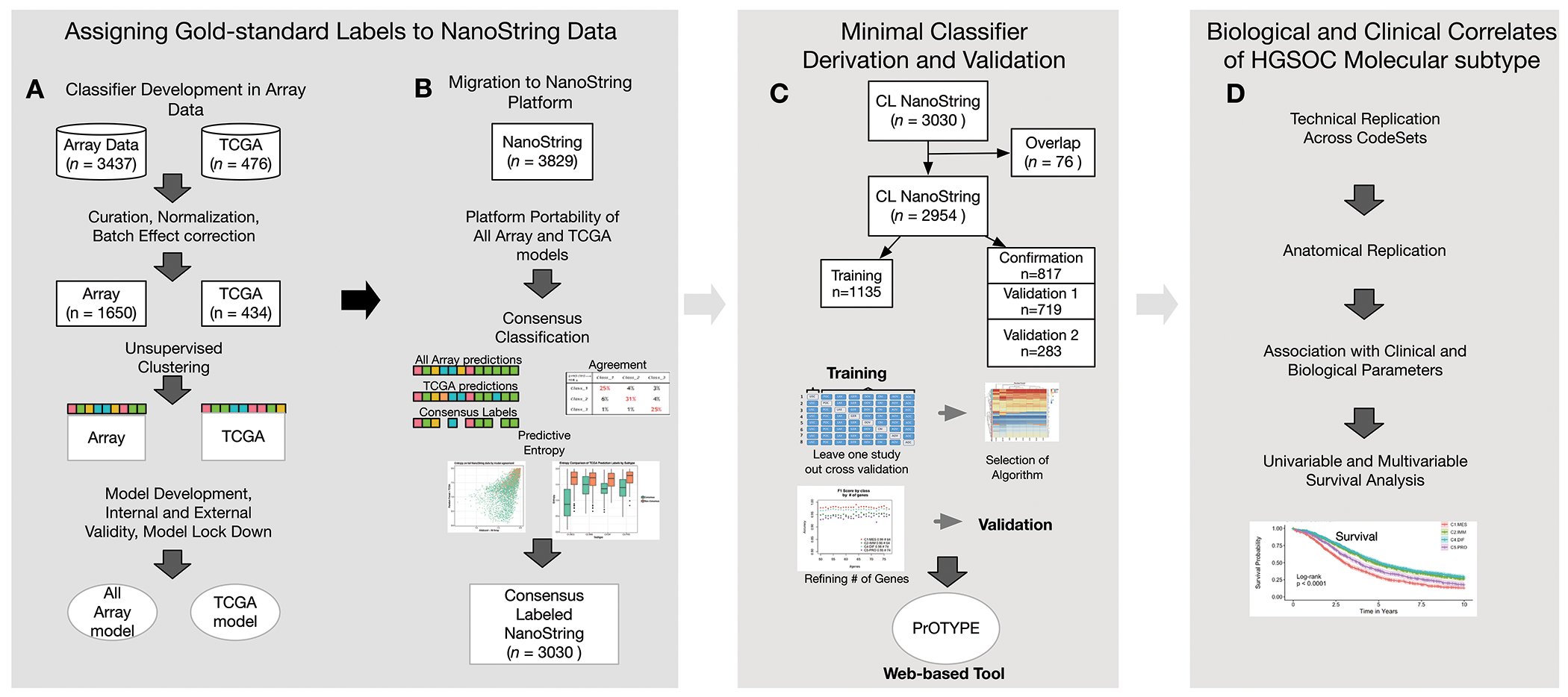
A schematic representation of the process we followed to obtain a final, clinical-grade classifier for HGSOC. Please note that the schematics above are for orientation only and are not intended to be interpreted. In the first panel we outline how de facto subtype labels were assigned to NanoString data, starting with (A) two parallel approaches to build models from array data, and (B) applying the resulting final models onto the NanoString dataset, where the consensus of the two methods became the de facto gold standard with 79% (n=3030) of our total NanoString cohort having agreement, consensus label (CL). In the second panel, (C) we provide the framework used to derive a minimal gene set classifier using the CL NanoString data after removing samples that overlapped both the NanoString and Array datasets (overlap n=76). Finally, in (D) a synopsis of the biological and clinical correlates that were investigated to confirm the biological validity of gene-expression based subtypes compared to previous work.
