Figure 4:
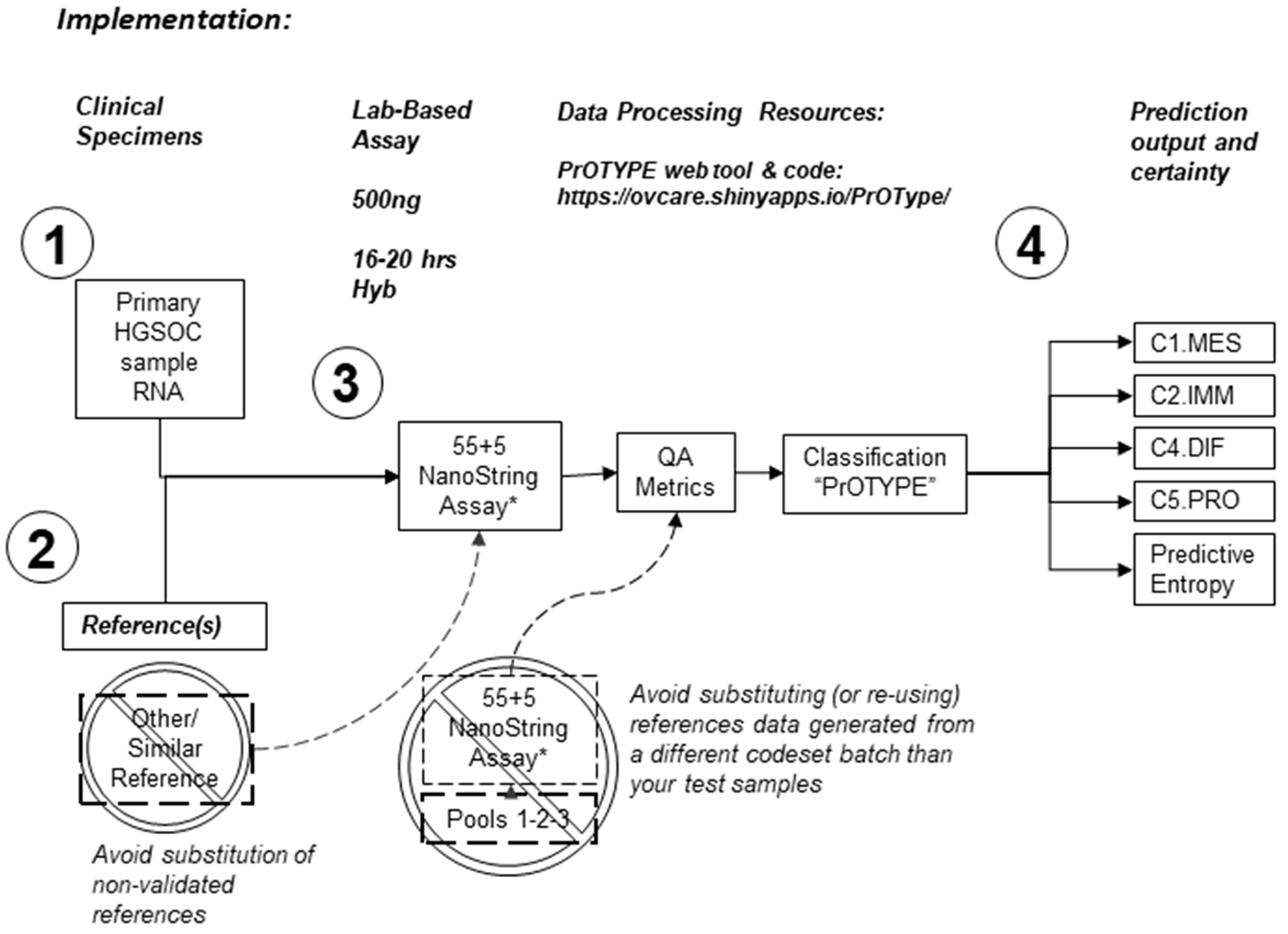
Locked-down Predictor of Ovarian carcinoma molecular subTYPE (PrOTYPE). The schematic illustrates the four critical components of the clinical-grade PrOTYPE assay (Supplement E) consisting of: (1) 500ng of total RNA from primary chemo-naïve HGSOC and (2) 100ng (each) of validated reference specimens. Each of these assayed individually by mixing specimen RNA with a (3) custom NanoString CodeSet (Supplemental Table SC7) containing 55 prediction model gene probes and 5 control gene probes (*55+5 NanoString Assay). RNA is hybridized with CodeSet, processed on a NanoString nCounter Prep-Station, and imaged at maximum fields of view on a NanoString nCounter Digital Analyzer. Resulting raw data is then normalized and HGSOC molecular subtypes predicted with our PrOTYPE computational algorithms using either a web-based tool, or R-script. This process will return (4) a prediction probability for the assayed specimen, for each subtype, and a single predictive entropy value. The latter can be used to estimate the certainty of prediction where 0 entropy corresponds to a near perfect prediction or “pure” subtype, while 2 entropy corresponds to near equal chance of assignment to any subtype.
