Figure 1. Molecular mechanism of lysosome positioning.
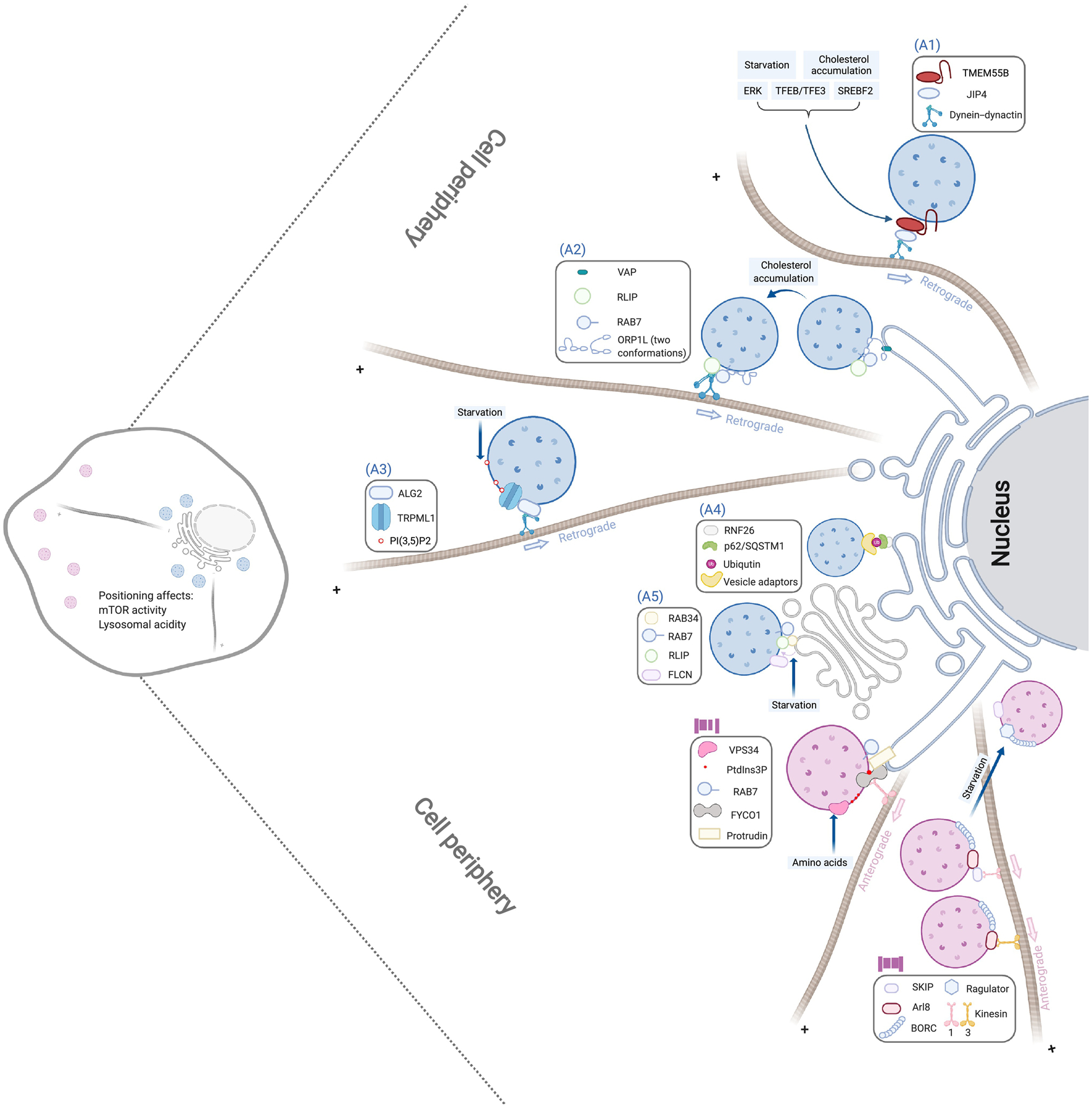
The perinuclear clustering of lysosomes (marked by blue) is driven by the following machineries: (A1) TMEM55B binding with the dynein adaptor JIP4 to recruit the dynein-dynactin complex; (A2) ORP1L adopting a conformation to facilitate the detachment of lysosomes from ER and the recruitment of dynein-dynactin by the Rab7-RILP complex; (A3) Lysosomal lipid PI(3,5)P2 activating the TRPML1 calcium channel to allow the recruitment of dynein-dynactin by the calcium sensor ALG2; (A4) ER-located RNF26 ubiquitinating p62/SQSTM1 and recruiting various vesicle adaptors through their ubiquitin-binding-domains; and (A5) Lysosome-associated FLCN promoting the interaction between lysosomal Rab7 and RILP and Golgi-localized Rab34. On the other hand, periphery positioning of lysosomes (marked in pink) is mediated by (B1) VPS34-induced PtdIns3P to promote the transfer of kinesin-1 from ER Protrudin to lysosomal FYCO1 and (B2) BORC recruitment of Arl8 to interact with different kinesins, which is blocked by its interaction with Ragulator. Each of these pathways can be regulated by different stimuli as indicated in light blue boxes, and lysosomal positioning regulates lysosomal acidity and mTOR activation. These pathways are shown as independent for presentation clarity, but multiple machineries can be present in a single lysosome.
