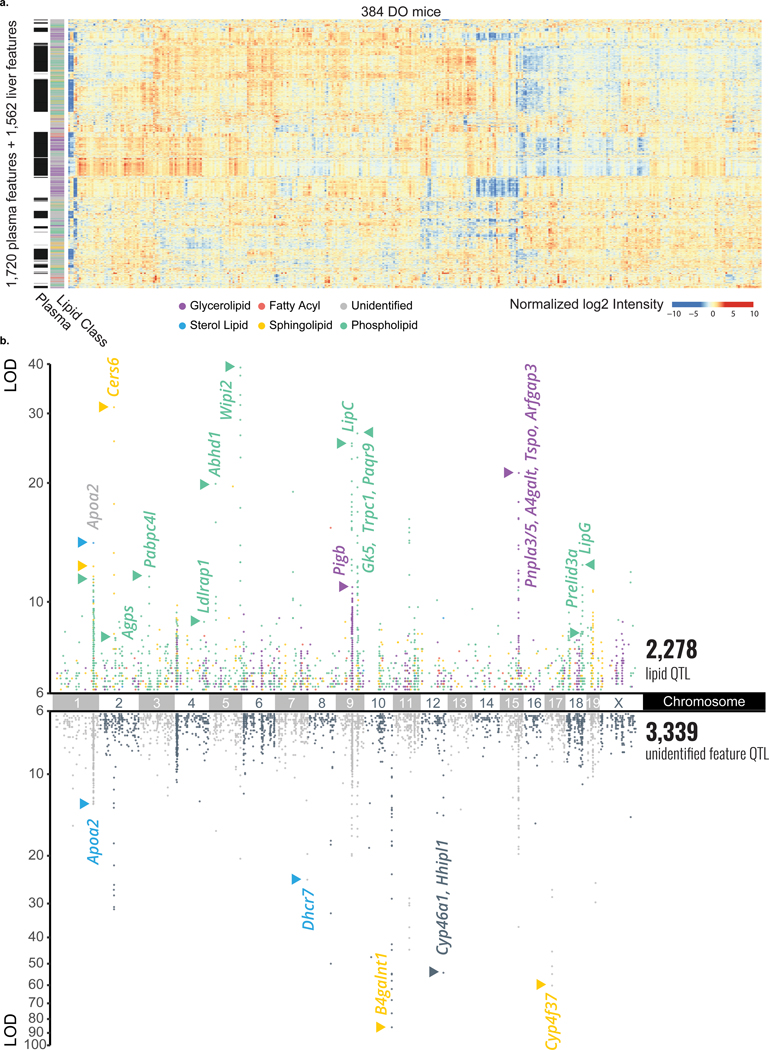
Figure 2. Large scale lipid quantitative profiling and subsequent QTL mapping reveals hotspots of associated lipids.
a, In plasma, we quantified 1,721 lipidomic features, 621 of which were identified, and in liver, we quantified 1,562 lipidomic features, 615 of which were identified. Hierarchical clustering of all 3,283 lipidomic features’ intensities by the 384 DO mice resulted in distinct clustering by lipid class, notably across tissue type. b, When mapped onto the mouse genome, 1,405 plasma and 1,190 liver features showed at least one QTL with an LOD > 6 as displayed in a Manhattan plot (n = 3,353 + 2,269 = 5,622 total QTL). A number of lipid hotspots are shared by identified lipids and unidentified features (e. g. at Apoa2), while others only appear among the unidentified features (e. g. at B4galnt1). Abbreviations: MTBE (methyl-tert-butyl ether), Chr (chromosome), DO (diversity outbred), ESI (electrospray ionization), LC-MS/MS (liquid chromatography - tandem mass spectrometry), QTL (quantitative trait loci), m/z (mass-to-charge), RT (retention time), LOD (logarithm of odds).