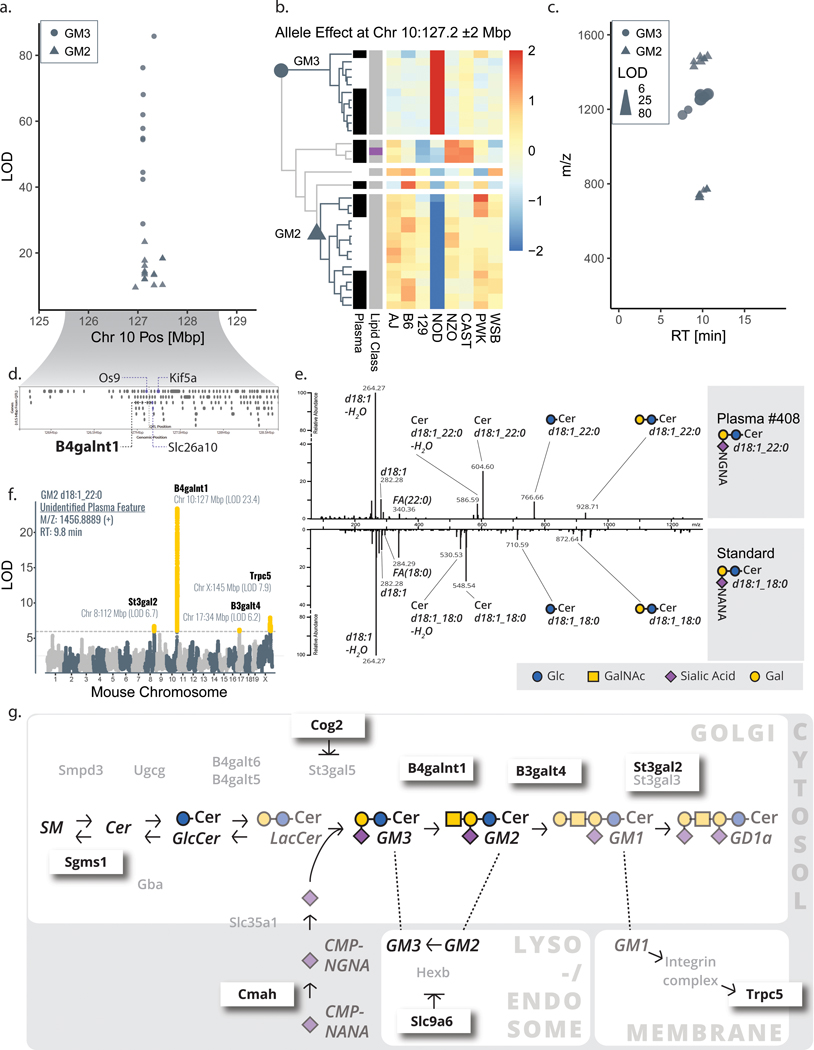
Figure 4. Lipid features mapping to B4galnt1 lead to identification of GM2 and GM3 gangliosides.
a, A hotspot of solely unidentified features with exceptionally strong correlation was composed of 11 liver and 15 plasma features mapping to chromosome 10:127 Mbp with b, a similar NOD-driver allele pattern (two main clusters from hierarchical clustering, row-scaled, Euclidean cutoff of h=2.5). Two groups of lipid features (circles vs. triangles) emerged as distinct in strength of LOD (a), directionality of allele effect (b), and m/z space (c). d, The candidate gene B4galnt1 pointed us to the putative identifications of GM3 (circles) and GM2 (triangles) gangliosides, which were confirmed by e, spectral matching with a human GM3 standard. f, Secondary QTL for these gangliosides, as exemplary shown for GM2 d18:1_22:0, mapped to eight additional candidate genes within 4 Mbp of the 15 total ganglioside hotpots that were previously linked to ganglioside metabolism. g, The various candidate genes influencing GM3 and GM2 levels span well-known enzymes (e. g. B3galt4) but also include indirect affectors including Cog2 and Slc9a6. Abbreviations: NOD (non-obese diabetic mouse strain NOD/ShiLtJ), Mbp (megabase pair), LOD (logarithm of odds), QTL (quantitative trait loci), Chr (chromosome), m/z (mass-to-charge), RT (retention time), Glc (glucose), GalNAc (N-acetylgalactosamine), Gal (galactose), SM (sphingomyelin), Cer (ceramide), NGNA (N-glycolylneuraminic acid), NANA (N-acetylneuraminic acid).