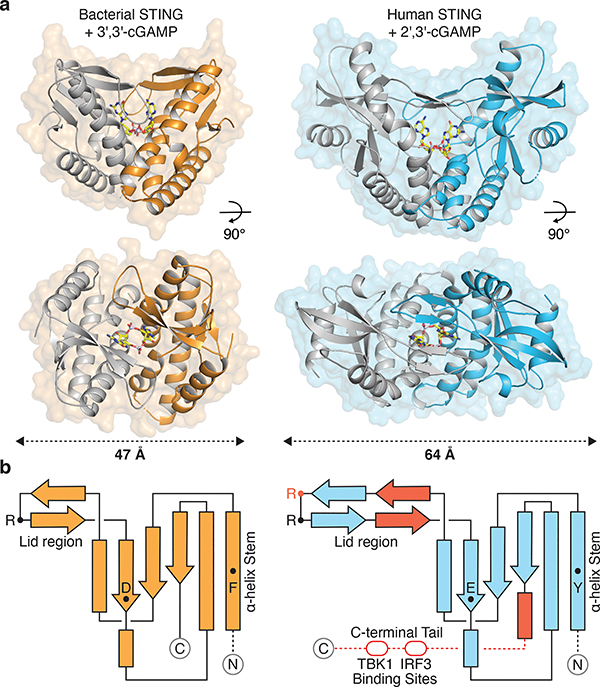
Figure 1 |. Structure of bacterial STING and definition of metazoan-specific insertions.
a, Crystal structure of a STING receptor from the bacterium Flavobacteriaceae sp. (orange) in complex with the cyclic dinucleotide 3′,3′-cGAMP. The FsSTING–3′,3′-cGAMP structure demonstrates a conserved mechanism of cyclic dinucleotide sensing shared between bacteria and human cells, and allows direct comparison with the human STING–2′,3′-cGAMP complex (PDB 4KSY, blue). For clarity, one monomer of each homodimer is depicted in grey.
b, STING topology diagrams denoting α-helices (rectangles), β-strands (arrows), and residues important for ligand recognition (FsSTING F92, D169, R153; hSTING Y167, E260, R232 in red, R238). Bacterial STING receptors reveal a minimal protein architecture required for cyclic dinucleotide sensing and allow direct definition of the structural insertions (red) in metazoan STING sequences required for signaling in animal cells.