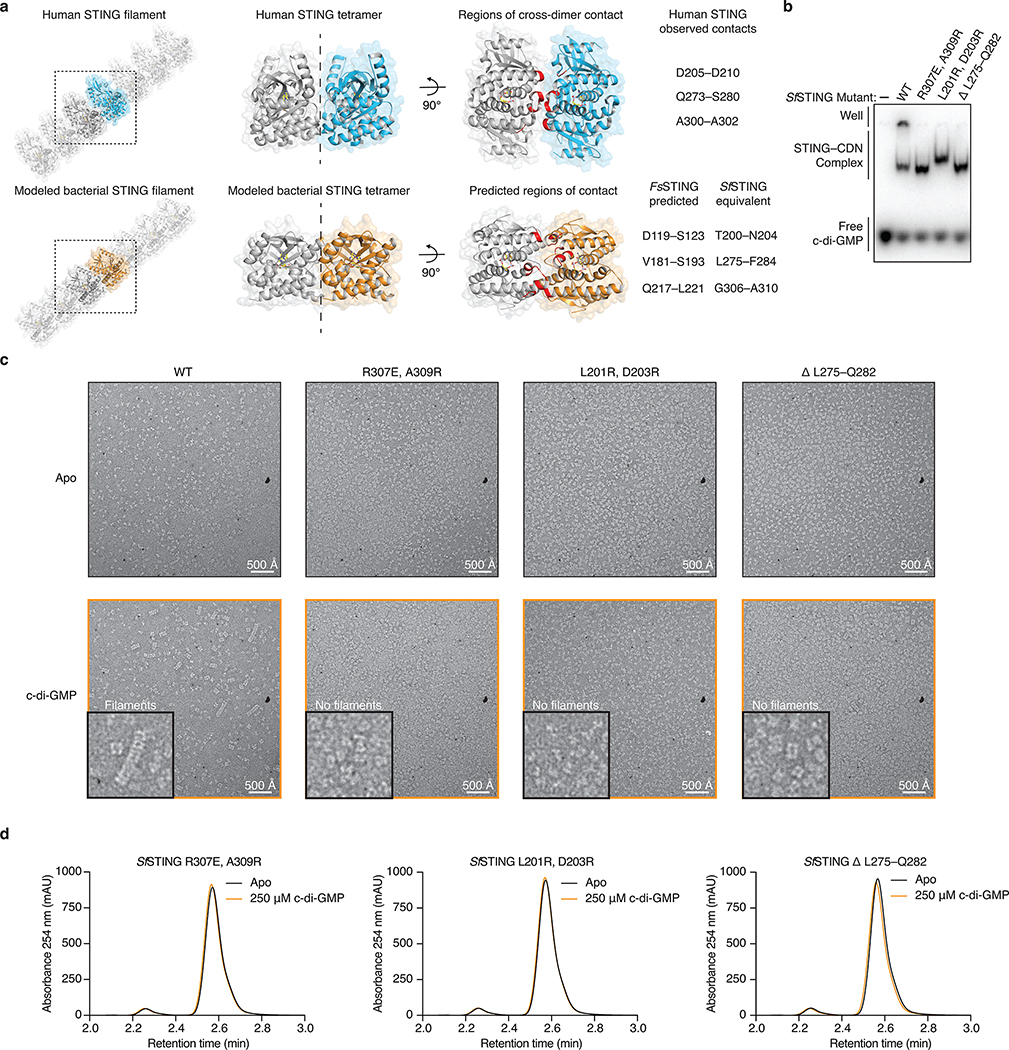
Extended Data Figure 8 |. Filament formation is required for bacterial TIR-STING activation.
a, Model of bacterial STING oligomerization and identification of surfaces involved in c-di-GMP-mediated filament formation. Electron microscopy analysis of SfSTING in the presence of c-di-GMP reveals filament formation likely occurs through parallel stacking of the homodimeric cyclic dinucleotide binding domain (see Extended Data Figure 7). To construct a potential model of this interaction, we used the X-ray crystal structure of the human STING–2′,3′-cGAMP complex (PDB 4KSY)7 and the cryo-electron microscopy structure of the chicken STING tetramer (PDB 6NT8)9 (top) as guides to position the FsSTING–3′,3′-cGAMP complex structure into a tetrameric conformation. The resulting model predicts that oligomerization-mediating surfaces in SfSTING include T200–N204, L275–F284, and G306–A310.
b, EMSA analysis of SfSTING variants indicates that mutations to the predicted oligomerization surfaces do not prevent c-di-GMP recognition. SfSTING mutants tested include R307E/A309R, L201R/D203R, and the loop L275–Q282 replaced with a short GlySer linker (GSGGS). Data are representative of 2 independent experiments.
c, Electron microscopy analysis of SfSTING variants in the presence of c-di-GMP. Mutations to the SfSTING surfaces identified in the structural model prevent all observable cyclic dinucleotide-induced filament formation supporting their predicted role in mediating oligomerization and bacterial STING filament formation. Images are each representative of n = 5 micrographs for each condition.
d, HPLC analysis of mutant SfSTING NAD+ cleavage activity in the presence of 500 nM protein and 250 μM c-di-GMP for 3 h. In the absence of cyclic dinucleotide-mediated oligomerization all SfSTING NADase activity is lost, confirming the requirement of filament formation in TIR domain activation. Data are representative of 3 independent experiments.