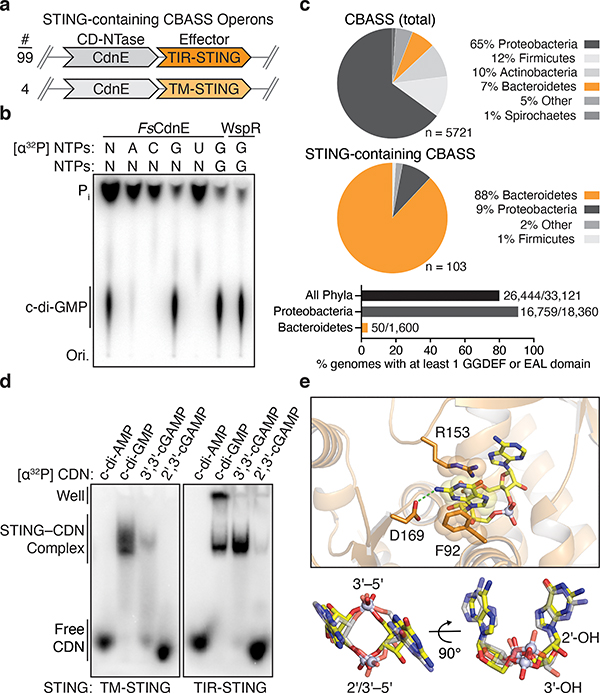
Figure 2 |. Bacterial STING systems signal through the 3′–5′-linked nucleotide second messenger c-di-GMP.
a, Schematic of STING-containing CBASS prokaryotic defense operons. STING is encoded downstream of a Clade E CD-NTase nucleotide second messenger synthase (CdnE) and exists as a fusion protein appended to Toll/Interleukin-1 receptor (TIR) or transmembrane (TM) effector modules with 99 and 4 sequences found, respectively.
b, Thin-layer chromatography analysis of Flavobacteriaceae sp. CdnE (FsCdnE) nucleotide second messenger synthesis. CdnE enzymes in STING-containing CBASS operons require GTP for catalysis and specifically synthesize the 3′–5′-linked product c-di-GMP. Control c-di-GMP synthesis reactions performed with the GGDEF enzyme P. aeruginosa WspR. N, all four radiolabeled NTPs; Pi, inorganic phosphate; Ori., origin. Data are representative of 3 independent experiments.
c, Analysis of the genomic context of all STING-containing CBASS operons. STING-containing CBASS operons in bacteria are occur mainly in Bacteroidetes (top). Sequenced Bacteroidetes genomes are generally devoid of canonical GGDEF and EAL c-di-GMP signaling domains (bottom) and 97 of 99 STING-containing bacteria lack any other predicted c-di-GMP signaling component. Loss of canonical c-di-GMP signaling provides an explanation for co-option of c-di-GMP as a CD-NTase immune signal. Numbers near bar graphs denote genomes containing at least one gene with a GGDEF or EAL domain, out of total genomes in the analyzed database.
d, Electrophoretic mobility shift assay monitoring bacterial STING–cyclic dinucleotide complex formation. Bacterial TM-STING and TIR-STING proteins preferentially recognize c-di-GMP and exhibit weaker affinity for 3′,3′-cGAMP. Bacterial STING receptors are unable to recognize the mammalian second messenger 2′,3′-cGAMP. TM-STING (R. ehrenbergii, ΔTM) and TIR-STING (S. faecium, full-length). Data are representative of 3 independent experiments.
e, Zoom-in cutaway of the cyclic dinucleotide binding pocket in the FsSTING–3′,3′-cGAMP structure. Top, FsSTING makes sequence-specific contacts to the guanine base consistent with preferential c-di-GMP recognition. Bottom, bacterial STING recognizes 3′,3′-cGAMP (yellow) in a compact conformation similar to 2′,3′-cGAMP (grey) in complex with human STING (PDB 4KSY).