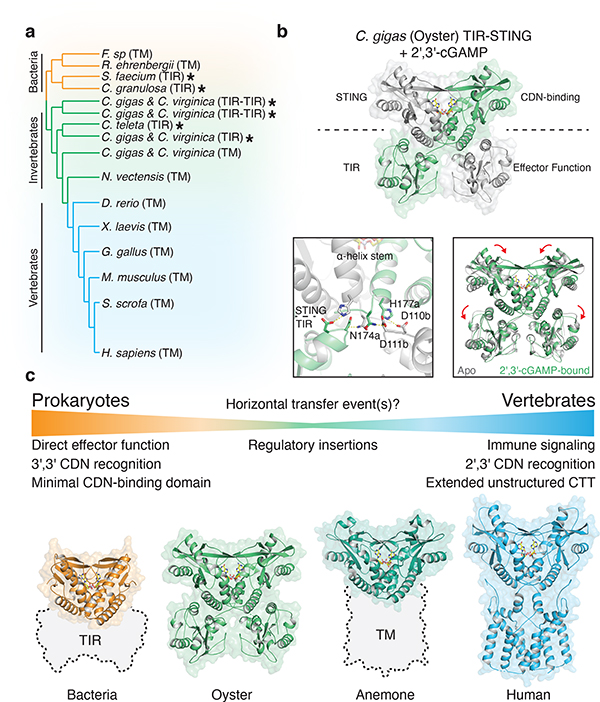
Figure 4 |. Structural basis of STING adaptation for signaling regulation.
a, Schematic derived from a structure-guided alignment of all known bacterial and metazoan STING sequences. Black stars denote TIR-STING fusions encoded in bacterial and metazoan genomes. Oysters encode several TIR-STING variants including genes predicted to contain multiple TIR domains (TIR–TIR).
b, Crystal structure of the full-length C. gigas (oyster) TIR-STING receptor (Genbank sequence XP_011430837.1) in complex with 2′,3′-cGAMP. Top, oyster TIR-STING adopts a domain-swapped conformation with TIR domains appended across a central dimeric axis. Bottom left, 2′,3′-cGAMP recognition results in new interactions between the STING domain α-helix stem of one monomer and the TIR domain of the other monomer. Bottom right, Ligand binding induces STING domain lid closure and a downward rotation that repositions the appended TIR domains.
c, Comparative structural analysis of STING adaptation suggests a model for the origin of cyclic dinucleotide sensing in innate immunity. STING evolved as a c-di-GMP sensor in prokaryotic bacteriophage defense, and acquisition in early metazoan cells may have allowed recognition of bacterial cyclic dinucleotides. Metazoan-specific structural insertions adapted STING for recognition of endogenous 2′,3′-cGAMP signaling and enabled antiviral and anti-tumor immune signaling in vertebrate cells. Structures shown: bacterial STING–3′,3′-cGAMP (FsSTING), oyster STING–2′,3′-cGAMP (C. gigas), sea anemone STING–2′,3′-cGAMP (N. vectensis PDB 5CFQ), human STING–2′,3′-cGAMP (H. sapiens PDB 6NT5 modeled with 6NT7).