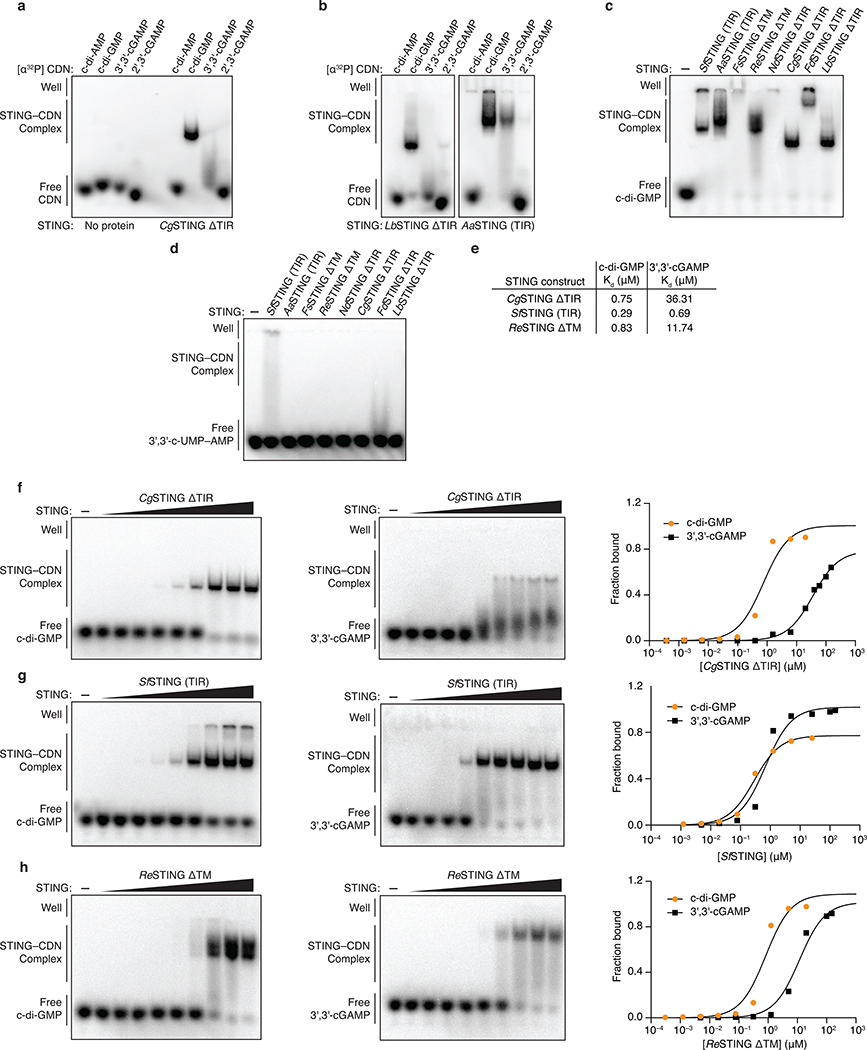
Extended Data Figure 4 |. Biochemical analysis of bacterial STING cyclic dinucleotide recognition specificity.
a,b, Electrophoretic mobility shift assay (EMSA) of purified bacterial STING proteins with radiolabeled cyclic dinucleotide (CDN) ligands. Bacterial STING receptors specifically recognize c-di-GMP and have a weak ability to bind 3′,3′-cGAMP. No interaction was observed with c-di-AMP or 2′,3′-cGAMP. Capnocytophaga granulosa (CgSTING), Lahnospiraceae bacterium STING (LbSTING), Aggregatibacter actinomyetemcomitans STING (AaSTING). Data are representative of 2 independent experiments.
c, EMSA analysis of a diverse panel of bacterial STING homologues demonstrates conservation of c-di-GMP binding in both TM-STING and TIR-STING CBASS immunity. NdSTING (Niabella drilacis) and FdSTING (Flavobacterium daejeonense). Higher-order complex formation visible as well-shifted complexes is consistent with STING oligomerization results (see Figure 3f and Extended Data Figure 7). Data are representative of 3 independent experiments.
d, EMSA analysis of diverse bacterial STING homologues broadly demonstrates no interaction with the 3′,3′-c-UMP–AMP second messenger synthesized by the divergent CD-NTase E. coli CdnE15 and further confirms the specificity of c-di-GMP signaling in bacterial STING-containing CBASS operons. Data are representative of 2 independent experiments.
e,f,g,h, EMSA analysis and quantification of the affinity of bacterial STING homologues for c-di-GMP and 3′,3′-cGAMP. Signal intensity analysis is plotted as fraction bound (shifted / total signal) as a function of increasing protein concentration and fit to a single binding isotherm. CgSTING and ReSTING have a >10-fold preference for c-di-GMP while SfSTING has a similar apparent affinity for c-di-GMP and 3′,3′-cGAMP. Data are representative of 2 independent experiments.