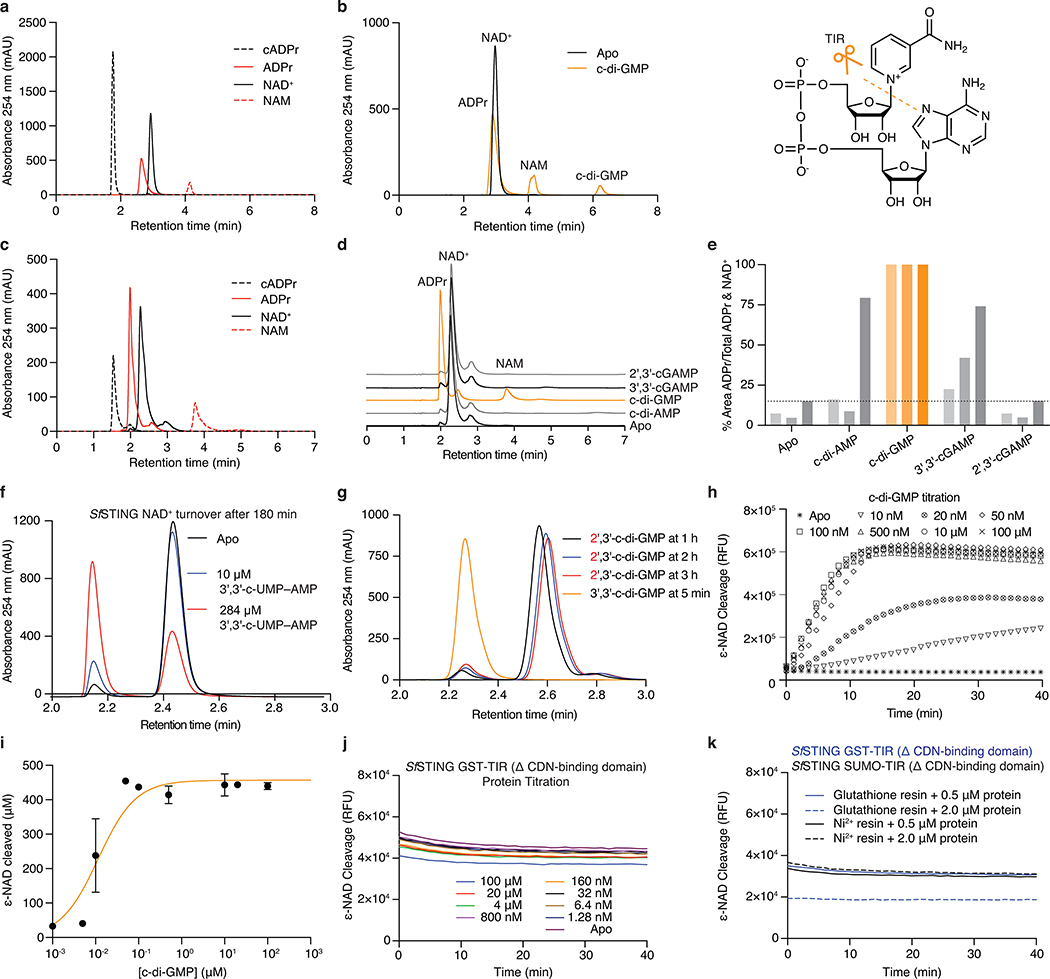
Extended Data Figure 5 |. Bacterial STING activation of TIR NADase activity.
a, HPLC analysis of chemical standards separated with an ammonium acetate:methanol gradient elution used to analyze bacterial TIR-STING activity (see Methods). The NAD+ and ADPr peaks have overlapping bases under these conditions. cADPr, cyclic adenosine diphosphate-ribose; ADPr, adenosine diphosphate-ribose; NAD+, β-nicotinamide adenine dinucleotide; NAM, nicotinamide.
b, HPLC analysis of SfSTING NAD+ cleavage activity with gradient elution. SfSTING at 500 nM protein with 2 μM c-di-GMP converts 500 μM NAD+ into ADPr and NAM in 30 min at ambient temperature. SfSTING does not generate any cyclized product and is therefore a standard glycosyl hydrolase. Inset: Schematic of NAD+ cleavage reaction.
c, HPLC analysis of chemical standards separated with an alternative isocratic elution strategy (see Methods) that results in clearer separation of NAD+ and ADPr peaks.
d, HPLC analysis of SfSTING NAD+ cleavage activity with isocratic elution. SfSTING NAD+ cleavage activity requires specific activation with c-di-GMP (30 min reactions).
e, HPLC analysis of SfSTING NAD+ cleavage activity and cyclic dinucleotide agonist specificity. Each reaction was tested with 500 nM SfSTING, 500 μM cyclic dinucleotide, and 500 μM NAD+ and sampled at 45, 90 or 180 min (gradient coloring in bars). SfSTING preferentially responds to c-di-GMP, but 3′,3′-cGAMP and c-di-AMP can function as weak agonists. Data are representative of 3 independent experiments.
f, HPLC analysis of SfSTING NAD+ cleavage activity in the presence of 3′,3′-c-UMP–AMP. 3′,3′-c-UMP–AMP is a >1000× weaker agonist than c-di-GMP. Data are representative of 3 independent experiments.
g, HPLC analysis of SfSTING NAD+ cleavage activity in the presence of a synthetic c-di-GMP analog with a noncanonical 2′–5′ linkage (2′,3′-c-di-GMP). 2′,3′-c-di-GMP is not capable of stimulating robust SfSTING activation even at very high concentrations (250 μM vs. 250 nM canonical c-di-GMP), confirming the specificity of bacterial STING for 3′–5′-linked cyclic dinucleotides. Data are representative of 3 independent experiments.
h, Plate reader analysis of SfSTING NAD+ cleavage activity using the fluorescent substrate ε-NAD. ε-NAD increases in fluorescence intensity after cleavage. SfSTING exhibits rapid catalysis with complete turnover at 500 nM protein with 500 nM c-di-GMP after 10 min at 25°C. No background activity is observed in the absence of ligand. Data are representative of 3 independent experiments.
i, Plate reader analysis of SfSTING NAD+ cleavage activity in the presence of 500 nM protein with increasing c-di-GMP concentration reveals that low nM c-di-GMP levels are sufficient to induce ε-NAD cleavage. Greater c-di-GMP concentrations are required for maximal activity consistent with binding data and the higher amount of c-di-GMP required for complete stabilization of the SfSTING–c-di-GMP complex (See Extended Data Figure 4e–h). Saturation occurs above 100 nM c-di-GMP with 40 min reactions. Data are ± s.d. of n = 3 technical replicates and are representative of 3 independent experiments.
j,k, TIR-domain NAD+ cleavage activity requires protein oligomerization. For other systems21,22, TIR activation has been observed at very high in vitro protein concentrations or in the presence of affinity resins as an artificial oligomerization-inducing matrix21,22. We used a GST-TIR construct to express the SfSTING TIR domain in absence of the STING cyclic dinucleotide binding domain and observed that even at >200× the concentrations for which the full-length protein shows c-di-GMP induced activity, or in the presence of multivalent affinity resin, no NAD+ cleavage activity occurs. These results demonstrate NAD+ cleavage activity specifically requires STING cyclic dinucleotide recognition for activation. Data are representative of 2 independent experiments.