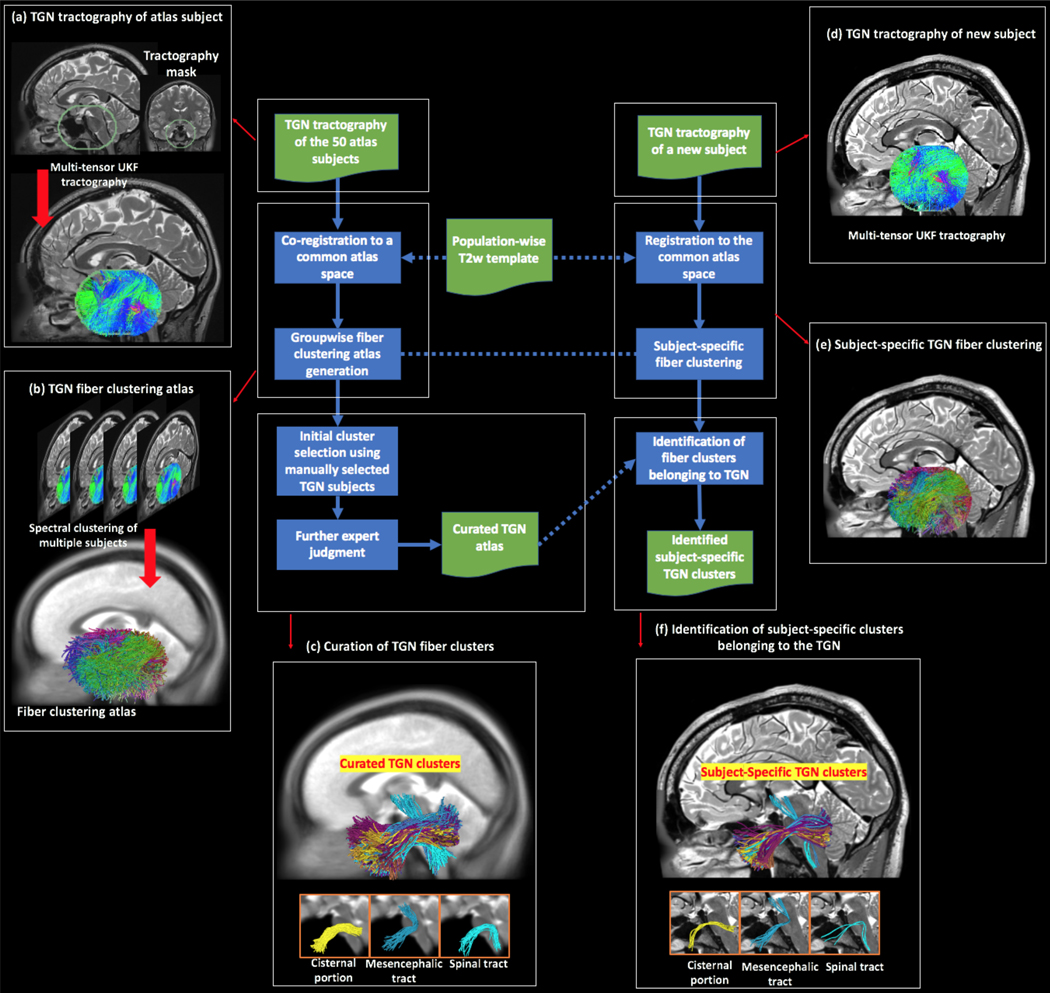
Figure 2.
Schematic diagram of the proposed method, where the blue boxes represent the computational steps and the green boxes represent the input/output. The major steps are linked to a pictorial illustration, as follows. (a) to (c) show the creation of the TGN atlas. (a) Multi-tensor tractography is seeded within a mask that covers the possible region through which the TGN passes. (b) Given the tractography data (co-registered to a common space, i.e., the atlas space) from the 50 atlas subjects, spectral clustering is performed to generate a fiber clustering atlas, where each cluster has a unique color. (c) Using expert neuroanatomical knowledge (involving three experts, GX, MAM, and NM), 40 clusters were identified to belong to the TGN, where each cluster represents a specific subdivision of the whole TGN. Three example clusters are displayed, belonging to the cisternal portion, the mesencephalic trigeminal tract, and the spinal trigeminal tract, respectively. (d) to (f) show identification of the TGN of a new subject. (d) TGN tractography of the new subject is performed, in the same way as the atlas subjects. (e) Fiber clustering of the tractography data (registered to the atlas space) is conducted according to the fiber clustering atlas. (f) Identification of the TGN clusters in the new subject is conducted by finding the corresponding subject-specific clusters to those annotated in the atlas. Three example subject-specific TGN clusters, corresponding to the ones shown in (c), are displayed.