Figure 7.
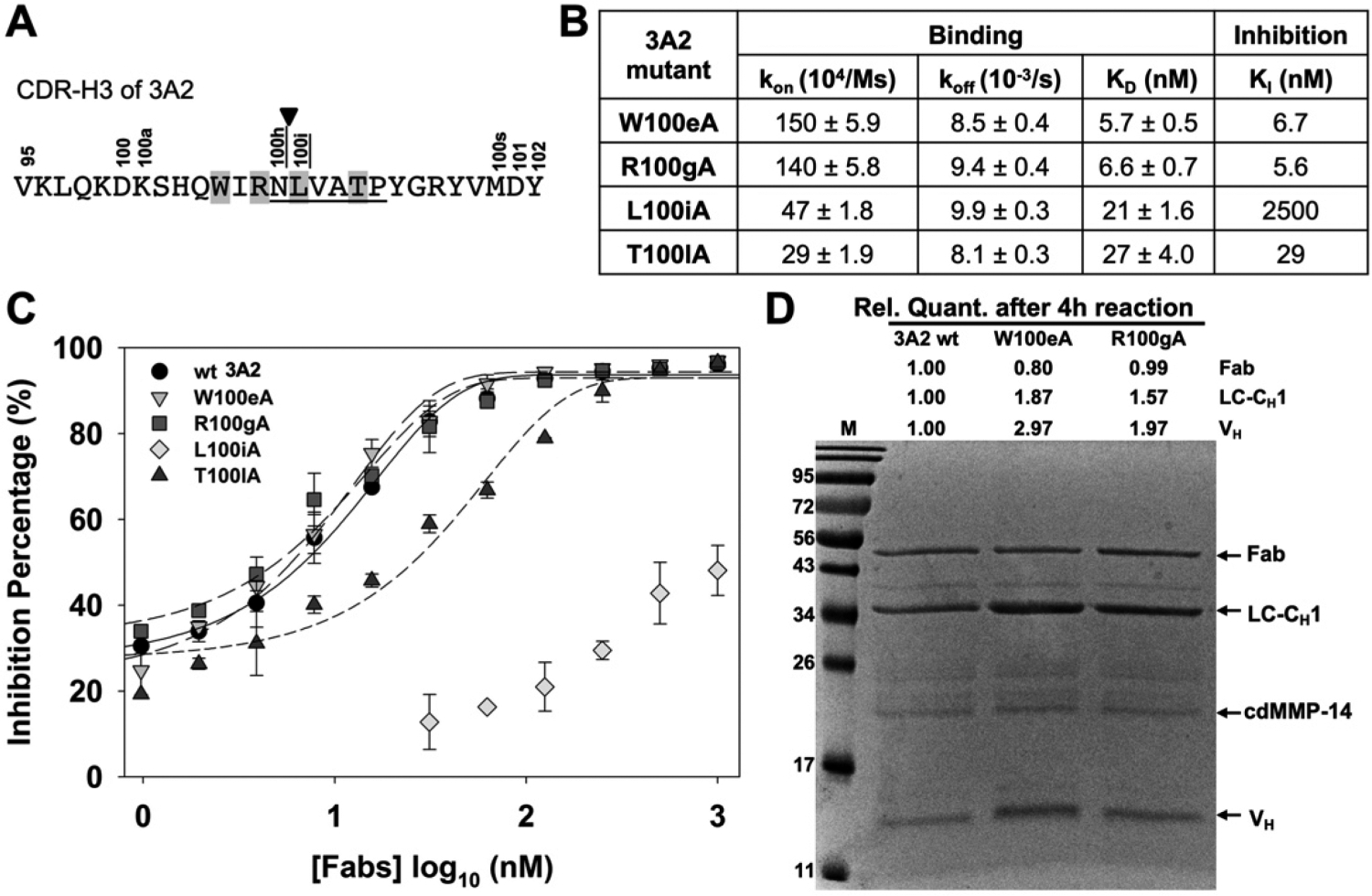
Characterizations of Fab 3A2 alanine point mutants. (A) CDR-H3 amino acid sequence of 3A2, showing the residues that match with the MMP-14 preferred substrate sequences (underlined, positions P1–P5′), cleavage site (arrowhead), and paratope positions chosen for alanine substitution (highlighted). (B) Binding kinetics and inhibitory potency data of 3A2 mutants. (C) Dose–response curves of inhibition measured with 30 nM cdMMP-14 and 1 μM FRET peptide substrate M-2350. (D) Fab stability assays with 3A2 wt and mutants W100eA and R100gA. Purified Fabs (2 μM) were incubated with 2 μM cdMMP-14 in 50 mM HEPES and 150 mM NaCl (pH 7.5) at 37 °C for 4 h and then analyzed by SDS–PAGE.
