Figure 1: Docking by MELD × MD successfully predicts bound structures in 23/30 cases.
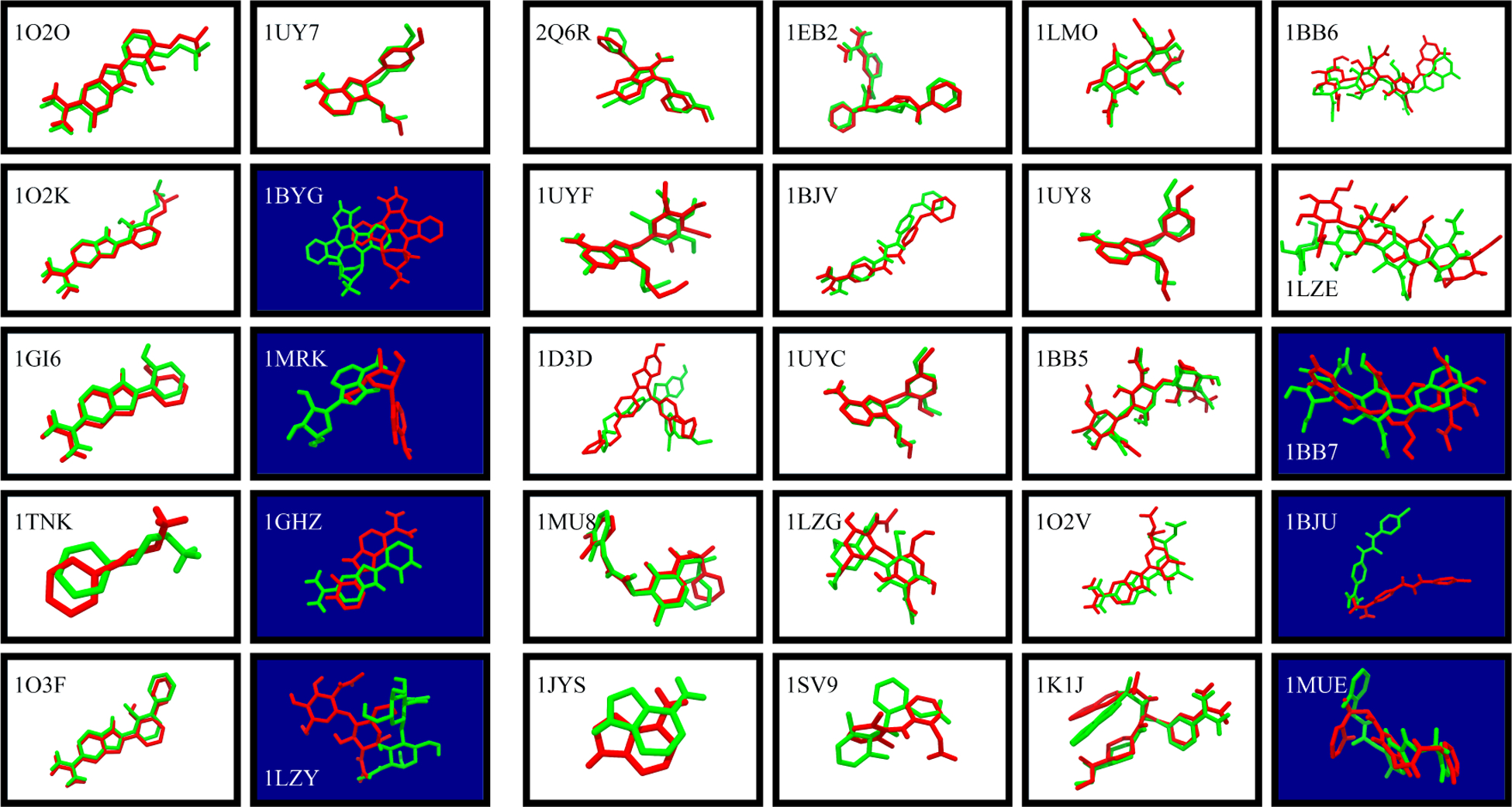
White panels show successful TOP1 predictions. Dark blue ones are not successes at the TOP1 level. (Green) Native ligand pose, superimposed on receptor Cα atom. (Red) MELD × MD predicted ligand poses. (Left 10 panels): DOCK by itself produces a native-like pose and rank it as TOP1, even without MELD × MD. (Right 20 panels): DOCK by itself fails to rank a native-like pose as TOP1.
