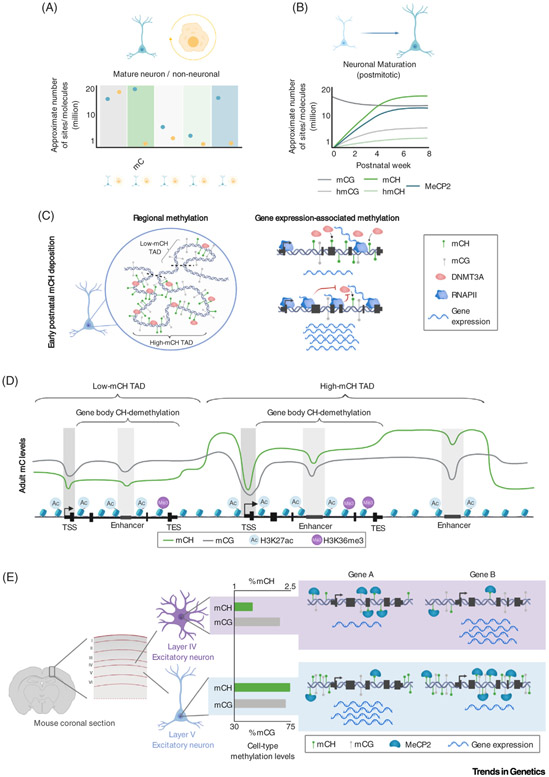
Figure 2. The Unique Neuronal Epigenome.
(A) Summary of reported levels of DNA modification sites and MeCP2 molecules between neurons and non-neural cell types, illustrating that mCH, hmCG, and MeCP2 are uniquely enriched within neurons. (B) During postnatal neuronal maturation, non-CG methylation, hydroxymethylation, and MeCP2 build up, reaching high levels in mature neurons at the young adult stage. (C) Left, a model depicting the early postnatal deposition of non-CG methylation by DNMT3A, establishing megabase-scale domains of high and low methylation associated with TADs. Right, a model illustrating how high levels of gene expression block DNMT3A activity, as highly expressed genes exhibit less DNMT3A binding and mCH accumulation compared with lowly expressed genes. (D) Illustration of typical methylation profiles in adult brain tissue. Both mCG and mCH are depleted at promoters and enhancers, but only mCH exhibits robust gene body demethylation and megabase-scale variations. H3K27ac and H3K36me3 histone modifications illustrate the relationship between DNA methylation and regulatory elements and transcriptional activity. (E) Illustration of differential global levels of mCH detected in subtypes of neurons. A layer V excitatory neuron shows substantially more mCH than a layer IV excitatory neuron [13]. These differences in methylation, combined with differential methylation of genes, set the stage for cell type-specific repressive effects by MeCP2 (e.g., gene A is more strongly expressed in layer V neurons than in layer IV neurons and gene B more so in layer IV than in layer V). Abbreviations: DNMT3A, DNA methyltransferase 3A; MeCP2, methyl CpG-binding protein 2; RNAPII, RNA polymerase II; TAD, topologically associating domain; TES, transcription end site; TSS, transcription start site.