Abstract
Breast cancer is the most common type of malignancy diagnosed in women. Through early detection and diagnosis, there is a great chance of recovery and thereby reduce the mortality rate. Many preliminary tests like non-invasive radiological diagnosis using ultrasound, mammography, and MRI are widely used for the diagnosis of breast cancer. However, histopathological analysis of breast biopsy specimen is inevitable and is considered to be the golden standard for the affirmation of cancer. With the advancements in the digital computing capabilities, memory capacity, and imaging modalities, the development of computer-aided powerful analytical techniques for histopathological data has increased dramatically. These automated techniques help to alleviate the laborious work of the pathologist and to improve the reproducibility and reliability of the interpretation. This paper reviews and summarizes digital image computational algorithms applied on histopathological breast cancer images for nuclear atypia scoring and explores the future possibilities. The algorithms for nuclear pleomorphism scoring of breast cancer can be widely grouped into two categories: handcrafted feature-based and learned feature-based. Handcrafted feature-based algorithms mainly include the computational steps like pre-processing the images, segmenting the nuclei, extracting unique features, feature selection, and machine learning–based classification. However, most of the recent algorithms are based on learned features, that extract high-level abstractions directly from the histopathological images utilizing deep learning techniques. In this paper, we discuss the various algorithms applied for the nuclear pleomorphism scoring of breast cancer, discourse the challenges to be dealt with, and outline the importance of benchmark datasets. A comparative analysis of some prominent works on breast cancer nuclear atypia scoring is done using a benchmark dataset which enables to quantitatively measure and compare the different features and algorithms used for breast cancer grading. Results show that improvements are still required, to have an automated cancer grading system suitable for clinical applications.
Keywords: Nuclear pleomorphism, Nuclear atypia scoring, Breast cancer, Histopathological image analysis
Introduction
Cancer is a group of different diseases in which the cells divide abnormally and tend to proliferate in an uncontrollable manner. If this proliferation is not controlled, it may result in the death of the patient. Though the reason for this abnormality is not exactly known even today, the established cause may be due to gene mutation of the DNA within the cells. Major causes for this gene mutation are smoking, carcinogenic chemicals, exposure to radiation, hormonal imbalances, etc. Cancer is of great concern today due to the rapid increase in the number of cancer patients. According to WHO, cancer is now the second leading death-causing disease globally, and has led to the death of around 8.8 millions in 2015. Nearly 1 in 6 deaths worldwide is due to cancer [20]. Mouth and prostate cancers are the most common malignancies among men whereas breast cancer is more prevalent among women with almost 25% of all cancers worldwide. According to [45], breast cancer ranks highest among Indian females with 25.8 per 100,000 females and mortality of 12.7 per 100,000 females.
Histopathologic Slide Preparation
The death rate owing to breast carcinoma can be considerably decreased by early and timely diagnosis and treatment. Advancements in the area of medical imaging techniques has brought about detection of breast cancer at the initial stages through proper screening before the symptoms appear. The most common diagnosis technique is to have a mammogram of the breast. But the mammography is not perfect. The rate of false positives and false negatives is high in mammography. Women with high risk of breast cancer are advised to have annual MRIs to be taken. If either mammography or an MRI shows any sign of having a disease, the only confirmation is through breast biopsies. There are different types of biopsies: either using some needle or through some surgical procedures. In a fine needle aspiration (FNA) biopsy, a very fine needle connected to a syringe is used to collect the sample for testing. In core needle biopsy, a large needle is utilized to collect the sample of the lesion. In certain cases, surgery is done to remove the lump for biopsy. The collected samples are then fed for processing, sectioning, and are placed on a glass slide and stained for the examination by an expert pathologist using a high-magnification microscope. This manual examination of the processed tissue under a microscope for a symptom of an ailment is termed as histopathology (or histology).
Immediately after the specimen is collected from the patient, it is subjected to a process known as fixation, which will preserve the tissue from the enzyme activity and prevent the decay [57] of the tissues. Commonly formaldehyde is used as the fixing agent, often called as “formalin.” After fixation, the specimen is grossed, processed, placed, and oriented in an embedding mold ready to be sectioned. Thin sections are cut out of the specimen using a special instrument known as “microtome” using fine steel blades. Usually the cells and other elements of the tissue are colorless and for tissue component visualization under the microscope; they are stained using one or more suitable stains to highlight the tissue structures. The most widely used stain is the H&E stains for providing distinguished structural information. In H&E staining, the tissue specimen is subjected to two stains: hematoxylin and eosin. Hematoxylin is a purple-blue dye which binds with the nuclear chromatin giving the nuclei a dark blue color and the eosin is an acidic pinkish dye which binds with the cytoplasm. A typical H&E stained breast biopsy tissue is shown in Fig. 1. The stained sections of the cancer tissues are covered using a glass coverslip and used for the histopathological analysis using microscope by an expert pathologist.
Fig. 1.

H&E stained breast biopsy tissue
Sometimes, H&E staining alone may not be giving a complete picture of the disease. In such cases, additional specialized staining techniques like immunohistochemical (IHC) staining may be required to gather more histological information. The IHC technique is mostly used for diagnosing malignancy of the tumor and for determining the stage of the tumor growth. IHC helps in determining the cells at the origin of a tumor through determining the absence or presence of some specific proteins in these observed sections of the tissue. Depending on the type of proteins detected by the IHC, specific and discrete therapeutic treatments are adapted for the detected cancer type. IHC often helps in identifying progesterone receptors (PR), human epidermal growth factor receptors 2 (Her2), or estrogen receptors (ER) which greatly affect the cancer proliferation [28, 74].
The visual analysis and grading of these specimens under a microscope by a pathologist is the widely accepted clinical standard for the detection and accurate diagnosis of breast tumors. The diagnosis of the disease is greatly influenced by the experience of the pathologist and is subjective, directly affecting critical diagnosis and treatment decisions. As there is a tremendous increase in the number of cancer patients, analyzing large number of slides manually is a highly laborious and time-consuming process for the pathologists. Also, this manual diagnosis often result in inter- and intra-observer variations, inconsistency, and lack of traceability. According to [64], there is a variability of 20% between experienced and novice pathologists in tumor diagnosis. A recent study on this diagnostic disagreement of pathologists in breast cancer diagnosis revealed that there is a disagreement of 75.3% [16] between individual and expert diagnosis. Thus, there is a great requisite for developing a computer-aided accurate cancer diagnosis and grading system that can overcome the problem of intra- and inter-observer inconsistency and thereby improvise the accuracy and consistency in the cancer detection and treatment planning processes.
Digital Histopathological Image Analysis
Automation of cancer diagnosis and grading require digitalization of the histological slides. This digitization of tissue slides is referred to as digital pathology, which could improve visualization, analysis of tissue slides, efficiency of pathological diagnosis, and treatment planning. Though the field of digital pathology originated in the 1980s, various factors like slow scanners, high cost, poor display mechanism, limited memory, and network have prevented it from being used in clinical diagnosis [36]. Digitalization of specimen slides were done in early days using digital cameras mounted on microscopes. The development of the whole-slide imaging scanner in late 1990s by Wetzel and Gilbertson [71] has brought about a turning point in digital breast histopathological image analysis. The scanning of conventional glass slides to produce digital slide, referred as whole-slide imaging (WSI), is currently used by pathologists worldwide. Some of the most commonly used WSI scanners include 3DHISTECH,1 Hamamatsu,2 GE Omnyx,3 Roche(previously Ventana),4 Philips,5 Leica(previously Aperio).6 These scanners are capable of performing scanning of the slides at a magnification of × 20 or × 40 times with 0.46 μ m/pixel or 0.23 μ m/pixel spatial resolution, respectively. The RGB images produced are usually compressed using JPEG or JPEG2000 standards, that provides multilayered storage of images, that enables fast panning and zooming.
The onset of whole-slide imaging has given a rapid advancement in the field of pathology, supported by high image quality, fast image acquisition techniques and increased storage capacity, and fast networking. With the advent of these WSI scanning techniques, it is now possible to automate breast cancer diagnosis using the digitized histopathological images and computer-aided diagnosis (CAD) methodologies [27]. CAD techniques have started been used for medical image analysis in early 1960s. But only during the last decade they are being widely used for histopathological image analysis. Now the interpretation of pathological images using CAD techniques has become a strong tool for exploring a wide range of breast histological image analysis tasks like (1) cancer detection, (2) grading of malignancy level of cancer or nuclear atypia scoring, (3) nuclei segmentation tasks, and (4) classification of cancer to various subtypes.
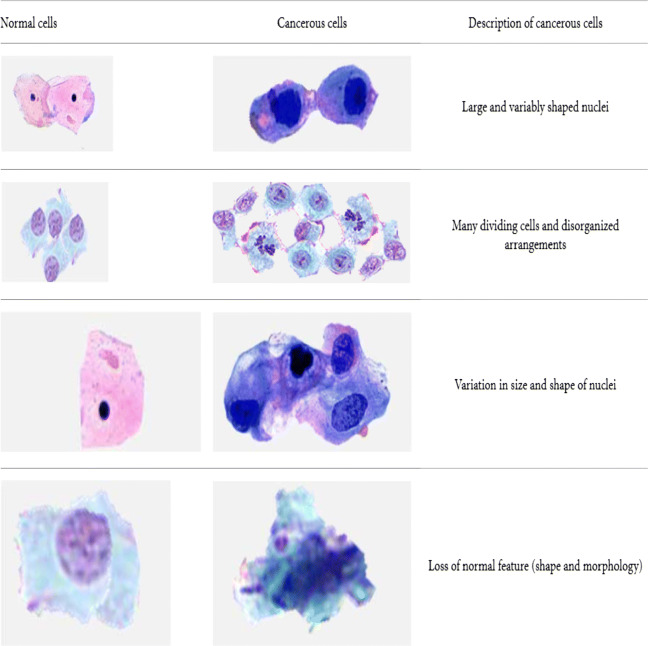
In histopathology, the cancer detection process normally consists of categorizing the image biopsy into cancerous one or non-cancerous one. The pathologists observe different characteristics like the shape, color, proportion of cytoplasm, and size of the cell nuclei and categorize the specimen. Figure 2 shows the difference between normal and cancerous cells [41]. The histological grading of cancer tissues, often called as nuclear atypia scoring, gives an estimate of patient prognosis and is helpful in developing patient-related treatment plans. The specimen is graded as low-grade, intermediate-grade, and high-risk breast cancer, according to the degree of tubular formation, nuclear pleomorphism, and mitotic activity. Automated nuclei segmentation and classification, often required for cancer detection and grading, is a recurring task and especially difficult in pathological images, since most of the nuclei appear in complex and irregular shapes and sizes. Studying the molecular level subtypes of breast cancer is often helpful in planning specific treatments and developing new therapeutic techniques. The profile regarding the cancer subtype can be determined through the genetic and molecular information obtained from tumor cells. Breast cancer includes mainly 4 significant molecular subtypes: Luminal A, Luminal B, HER2, and Triple negative/basal-like types.
Fig. 2.
Difference between normal and cancerous cells [41]
This review paper will be mainly focusing on nuclear atypia scoring aspect of breast carcinoma. The paper explores the different techniques and methodologies used for the grading of breast cancer, addresses the challenges involved, and discusses the strategies used by the image analysis techniques in overcoming these challenges. The study is a venture to abstract out the recent developments in breast cancer grading that sparks light into how knowledge has evolved within the field, spotlighting what has already been done, what is conventionally accepted, what is emerging, and what is the present status of research in this field. This helps in identifying the research gap, i.e, under-researched or unexplored areas that needs to be imbibed with research works in the right direction. The paper also gives an overview of the various evaluation metrics used for the quantitative analysis of nuclear pleomorphism scoring. The paper is organized as follows: the “Histological Grading of Breast Cancer Tissues” section will be giving an overview and challenges in nuclear atypia scoring, the “??” section will be dealing with the algorithms used for breast histological image grading, the “Evaluation Metrics” section summarizes the main evaluation metrics used for the evaluative analysis of cancer grading, and the “??” section will be dealing with the comparative analysis of major nuclear atypia scoring algorithms. A look into the future prospects of breast histopathological image analysis is given in the “Nuclear Atypia Scoring: Future Perspective” section and finally conclusions are drawn in the “Conclusion” section.
Histological Grading of Breast Cancer Tissues
Histological grading of cancer is the representation of a tumor based on how far the tumor tissues differ from normal tissue. Within the last decade, grading of hematoxylin-eosin (HE) stained histopathology image is often accepted as a standard practice for breast cancer prediction and prognosis. It provides an inexpensive and prognostic information about the biological characteristics and clinical behaviors of BC. Breast contains well-differentiated cells, that take specific shapes and structure based on their function within the organ, whereas cancerous cells lose this differentiation when compared with the normal cells. In an infected breast, the cells become disorganized, less uniform, and cell division occurs in an uncontrollable manner. Pathologists categorize the cancer as low-risk, intermediate-risk, or high-risk cancer depending on whether the cells are highly differentiated, moderately differentiated, or less differentiated, respectively, as the cell may progressively lose their characteristics observed in normal breast cells. Undifferentiated or poorly differentiated tumors may probably be growing and spreading at a faster rate with low survival rate.
Invasive breast cancers often spread out of the original site (either the lobules or milk ducts) into the neighboring breast tissues and they constitute almost 70% of all breast cancer cases [30], and they are usually having poor prognosis compared with the in situ subtypes. Further analysis on tumor differentiation can be done on isolation of invasive breast cancer. The cancer is graded depending upon different factors like the type of cancer. The internationally and widely recognized Nottingham grading system (NGS), an improved Scarf-Bloom-Richardson grading system, is the most widely used system for nuclear atypia scoring [23]. NGS forms a qualitative evaluation method where the atypia score is evaluated based on three morphological factors: extent of normal tubule structures, nuclear atypia or nuclear pleomorphism, and the count of mitotic cells [17].
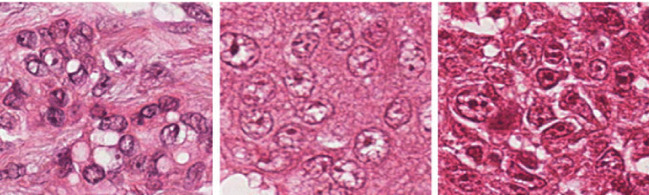
When evaluating tubules, the percentage of tumor that displays tubular structure is assessed. A score of 1 is given if the portion of area that is composed of definite tubules is more than 75% of the tumor area. If the area of tubule formation is between the values of 10 and 75%, a score of 2 is assigned and a score of 3 if the tubule formation area is less than 10%. In nuclear pleomorphism, a score of 1 is assigned when the nuclei are small in size, with regular outline and having a uniform distribution of nuclear chromatin. For cells with open, vesicular nuclei that shows visible nucleoli and with moderate variability in both size and shape, a score of 2 is given. Cells with prominent multiple nucleoli and vesicular nuclear structure is assigned a score of 3. Mitotic count determines the number of mitotic cells found in 10 High Power Fields (HPF) microscopes. If the number of mitotic cells are more, it implies a high-grade cancer. A score of 1 is given for up to 9 mitoses per 10 fields, score of 2 if there are 10 to 19 mitoses, and a score of 3 is given if more than 20 mitoses are observed. Table 1 gives a summary of Nottingham grading system (NGS) used for breast cancer scoring. The score from all three features are added up to get the overall breast cancer grade. If the total is between 3 and 5, it indicates that the tumor is well-differentiated and it is considered a grade I tumor. If the cells are moderately differentiated, the total score will be between 6 and 7, and hence treated as grade II cancer. Grade III cancers will be poorly differentiated with a total score between 8 and 9. Thus, the cancer will be graded with scores 1, 2, or 3, for the low, moderate or strong nuclear pleomorphism respectively. Sample images having a nuclear atypia score of 1, 2, and 3 [76] are given in Fig. 3.
Table 1.
Summary of Nottingham grading system (NGS) for breast cancer
| Feature | Score | Description |
|---|---|---|
| Tubule formation | 1 | ≥ 75% of the tumor forms tubule |
| 2 | 10–75% of the tumor forms tubule | |
| 3 | Less than 10% of the tumor forms tubule | |
| Nuclear atypia | 1 | Small, uniform, and regular nuclei |
| 2 | Moderate variations in size and shape | |
| 3 | Multiple nucleoli with prominent variation | |
| Mitosis count | 1 | 0–9 mitotic cells in 10 HPF |
| 2 | 10–19 mitotic cells in 10 HPF | |
| 3 | Greater than 20 mitotic cells in 10 HPF |
Fig. 3.
Sample images with NAS of 1, 2, and 3, respectively [76]
The currently existing manual visual analysis of the histopathological slides are highly subjected to the individual opinions of the pathologist and differ in intra- and inter-observer decisions, which greatly affects the disease diagnosis and the treatment prescribed. Hence, its highly essential to develop an automated cancer grading system based on quantitative digital image analysis techniques that can circumvent the observer variabilities and thus develop a consistent system for breast cancer diagnosis. Moreover, the quantitative analysis of histopathological digital images is not only essential for clinical applications but also for research purposes which may help us understand the biological mechanism and genetic abnormalities associated with the disease.
Challenges in Histopathological Image Analysis
For the past two decades, tremendous researches are going on computer-aided cancer diagnosis. Automated CAD analysis of digital histological images can ensure objectivity and reproducibility using digital image analysis techniques. Through image analysis techniques, various quantitative informations like size, shape, and deformities of the cells can be extracted. The major steps involved in digital image analysis of histopathological images include pre-processing, segmentation, feature extraction, feature selection, and classification. Many efficient computer-aided image processing algorithms are available for automated image analysis.
Automated cancer diagnosis can be considered a great promise for advanced cancer treatments, but it is not a forthright task, as a number of challenges need to be overcome. Especially, the breast histopathological image analysis has been a challenging job due to the numerous variabilities and artifacts instigated at slide preparation and also due to the complex structure of the cancer tissue architecture. The image analysis algorithms are greatly dependent on the image quality of the digital slides. Artifacts may be introduced in the stained slides due to many reasons like improper fixation, type of fixative used, errors in autofocusing, poor dehydration, or due to uneven microtome sectioning. The quality of the slides need to be ensured in terms of avoiding chatter artifacts and tissue folds induced during erroneous microtomic sectioning. Proper coverslipping has to be done to avoid creation of air bubbles and uniform section thickness has to be ensured [24].
Histological image analysis is also challenged, by improper staining and variations in lightning and scanning conditions leading to blurring, noise, and unwanted artifacts in the captured images. Color variations may occur in the tissue appearance, due to various reasons like difference in specimen preparation process, stain variations from different manufacturers or batches, usage of WSI scanners from different vendors, and difference in storage time of the stained specimen. The requirement for standardization of methods and reagents used in histological staining is discussed by Lyon et. al in [43]. Also uneven distribution of stain within the specimen tissue due to difference in concentration and timing create issues in processing the stained material. In the case of automated image analysis, this has been of more concern and hence before the image analysis, the images need to be normalized to reduce the effect of these staining variations. Stain normalization and color separation were used on the H&E stained images for the first time in [51]. Later on many explicit stain or color normalization algorithms have been applied as a pre-processing step in various algorithms for histopathological image analysis.
At the time of digital scanning of the slides, uniform light spectrum is used for illumination. Tissue auto-fluorescence (AF) variations in microscopic setup and differences in staining and sample thickness may cause uneven illumination across the tissue samples. Also the sensitivity of scanners is different for different wavelengths of the illuminating light. Usually the cameras exhibit low response for short wavelength signals like blue and show a high response at long wavelengths like red signals. These differences in the illumination need to be addressed before applying the image analysis techniques.
For feature extraction, mostly the nuclear structure of the tissue need to be segmented before extracting and selecting the suitable features. This nuclei or cell segmentation associated with cancer grading is another major challenge because of the complex structure and nature of the tissue specimen. This is of major concern in the high-grade tumors, where cells are poorly differentiated and nuclei are often hollow with broken membrane [33]. Segmentation is a challenging task in cases of specimen with occlusion, touching or overlapping clustered cells and tissues, which significantly influences the accuracy of nuclear pleomorphism scoring and cancer diagnosis.
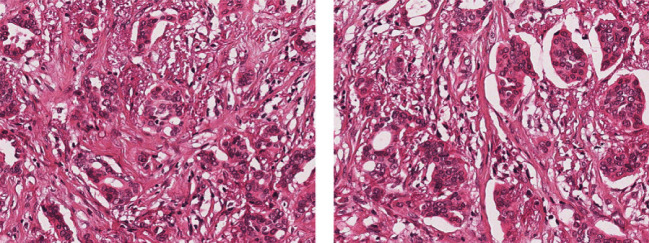
Feature selection refers to extracting and selecting relevant and important features from the histological images. This has now been an important area of research with the advancement of histopathological image analysis. It needs to represent properties of the tumor cells or tissues in a quantifiable manner. The features selected should be unique and distinguishable enough to automatically identify cancerous and non-cancerous tissues and to grade them accordingly. This is a challenging task, as in most cases, the overall appearance of images are quite similar, that it is quite difficult to quantify their properties using distinguishable extracted features. Figure 4 represents two sample images which are quite similar with respect to their appearance and texture but graded with NA score of 1 and 2.
Fig. 4.
Sample images with quite similar attributes and appearances, but scored with NAS of 1 and 2, respectively [40]
Last important challenge for the histopathological image analysis is the system evaluation. Due to the limitations in the availability of data, there may be chances of substantial amount of bias if the evaluation of the system is not done properly. Some algorithms may be claiming good results on limited dataset, but proper evaluation of these techniques can be done only if standardized and large collection of dataset are tested and assessed on them. This lack of unified benchmark dataset forms another major challenge, as most of the automated cancer diagnosis methods are carried out using their own private datasets, using different evaluation methods and diverse performance metrics. For the numerical comparison of these methods, benchmarked dataset is highly essential. This problem has been addressed to some extent with a few open scientific grand challenges conducted in the field of pathology images. The availability of standardized and annotated dataset in these challenges gives an opportunity of testing and evaluating different histological image analysis methods on the same data. This helps to have an objective comparison of the strengths and limitations of these methods. Some of the grand challenges conducted in the field of breast histopathological image analysis are discussed in the “??” section.
Grand Challenges in Breast Histopathological Image Analysis
Some of the challenges conducted in the histological image analysis of breast tumor includes AMIDA13, MITOS12, MITOS-ATYPIA14,7 TUPAC16,8 and BACH 2018 (BreAst Cancer Histology).9
The MITOS contest [58] for the detection of mitosis in H&E stained slides images of breast cancer was conducted in 2012 in connection with the conference ICPR 2012. Mitosis detection is a difficult task as often mitosis are small in size with large variations in shapes and the count of mitotic cells is considered a significant parameter for the accurate prediction of breast cancer outcome. The MITOS benchmark consists of 50HPFs from 5 different slides scanned at × 40 magnification. Seventeen teams participated in the contest and the performance of the best team was with an F1-score of 0.78. But the dataset provided was found to be too small to obtain a good assessment of robustness and reliability of the proffered algorithms.
Assessment of Mitosis Detection Algorithms 2013(AMIDA13) challenge was conducted in 2013 as part of MICCAI 2013 conference. AMIDA [68] benchmark re-edited the MITOS12 dataset with 12 training samples and 11 for testing, with more than one thousand mitotic figures annotated by multiple observers. A total of 14 teams submitted their methods with the highest F1-score of 0.611, which indicates progress still need to be made to reach clinically tolerable results.
MITOS-ATYPIA14 challenge at ICPR 2014 conference enlarged their MITOS2012 dataset and the challenge consists of two tasks: mitosis detection and evaluation of nuclear atypia score. The benchmark dataset includes hematoxylin and eosin (H&E) stained slides scanned by two WSI slide scanners: Aperio Scanscope XT and Hamamatsu Nanozoomer 2.0-HT. Several frames with × 20 magnification identified within the tumors in each slide are selected by experienced pathologists. These × 20 frames are considered for nuclear atypia scoring and they are further magnified at × 40 to obtain four subdivided frames. The × 40 frames are subjected to mitosis annotation and a scoring is performed for the six criteria related to nuclear atypia. The training data set consists of 284 frames at × 20 magnification and around 1136 frames at × 20 magnification. About 17 teams participated in the contest and the highest rank for the mitosis detection was for the algorithm with F1-score of 0.356. For nuclear atypia scoring contest, the highest secured point was 71.
As part of the MICCAI Grand Challenge, the Tumor Proliferation Assessment Challenge 2016 (TUPAC2016) was organized, which consisted of mainly three tasks: (1) Prediction of proliferation score based on mitosis counting, 2) Prediction of proliferation score based on molecular data, and 3) Mitosis detection. The training dataset consists of 500 breast cancer cases, represented with one whole-slide image and is annotated with a proliferation score based on mitosis counting by pathologists, and a molecular proliferation score. The highest score for the first task was with a quadratic weighted Cohen’s kappa score of 0.567, whereas for the second task, the highest Spearman’s correlation coefficient score was 0.617. For the mitosis detection task, a highest F1-score of 0.652 was secured.
Following the availability of these benchmark dataset, many histological image analysis techniques were proposed by researchers for breast mitosis detection, nuclear atypia scoring, and for cancer prognosis. But all these breast cancer diagnosis and prognosis methods are carried out on a small dataset as there was a shortage of public dataset containing large quantity of images. This problem was mitigated with the availability of the publicly available dataset, the BREAKHIS DATASET10 compiled by Spanhol et al. [61]. The Breast Cancer Histopathological Image Classification (BreakHis) consists of 9109 WSI images of breast cancer tissues obtained from 82 patients available at × 40, × 100, × 200, and × 400 magnifications. The availability of this dataset provides scope for the scientific community to have benchmarking and standardized evaluation to be made in this clinical area.
The recent BACH 2018 challenge conducted as part of the International Conference on Image Analysis and Recognition (ICIAR 2018) involves the task of classifying the histology breast cancer images into benign, normal, invasive carcinoma and in situ carcinoma. The dataset consist of high-resolution (2048 × 1536 pixels) images annotated by two experts and those images with difference of opinion are discarded. The highest overall prediction accuracy of 0.87 was achieved in the contest.
The most recently published BreCaHAD 2019 (breast cancer histopathological annotation and diagnosis) dataset [1] consists of 162 breast cancer histopathology images, that classify H&E stained images into six classes, i.e, mitosis, apoptosis, tubule, non-tubule, tumor nuclei, and non-tumor nuclei.
Though many datasets like TUPAC 2016, BREAKHIS, BACH 2018, and BreCaHAD 2019 challenge datasets related to breast cancer histopathological image analysis have been released, none of them consists of labeled samples for nuclear atypia scoring of breast cancer. In this regard, the MITOS-ATYPIA14 challenge dataset is the most recent one related to our problem of concern. Hence, we have adopted them for our comparative analysis.
In this paper, we present a systematic review of the computational steps involved in the nuclear atypia scoring or cancer grading on breast histopathological whole-slide images. In the following section, we will be explaining the different techniques and methodologies used for the grading of breast cancer, addressing the challenges involved and discussing the strategies used by these techniques in overcoming the challenges.
Histological Grading of Breast Cancer or Nuclear Atypia Scoring: Current Status
Grading of histopathological images of breast biopsy specimen is currently considered to be the golden clinical standard for the diagnosis and prognosis of breast tumor malignancy. Nuclear Atypia Scoring (NAS) is used as a quantitative diagnostic measure to assess the grade of different cancers, especially breast cancer. It provides a measure of the degree of variation in the shape and size of cancer nuclei as compared with normal nuclei in the breast. This histological grading along with other factors is used for the prognosis [9] and prediction of the disease progression, which help in choosing the best treatment method.
In the case of breast cancer, the visual examination of histological slides of biopsy specimen stained using hematoxylin and eosin (H&E) continues to be the standard procedure for cancer detection and determination of malignancy grade. Nottingham grading system (NGS), recommended by the World Health Organization (WHO), assesses the morphological measurement of tubule, mitotic count, and nuclear atypia for the breast cancer grading. Often these manual diagnosis require extensive training and experience for the pathologist to be a proficient in the technique and even then there will be great extent of disagreement between pathologists regarding the grade. This is particularly true in the case of nuclear pleomorphism, which is an indication of the size, shape, and chromatin distribution of the nuclei. This subjectivity of measurement and poor reproducibility have resulted in the great demand for an automated grading system. The advancements in the field of digitization of whole-slide histopathological images (WSI), increasing computing power and storage capacity, have made it practically possible to have histological breast cancer grading to be done fully digitally.
Histopathological image analysis algorithms for nuclear pleomorphism scoring can be broadly classified into two categories: handcrafted feature-based and learned feature-based algorithms. The handcrafted feature-based algorithms require image features to be explicitly extracted and mainly consists of image pre-processing, nuclei segmentation, feature extraction, feature selection, and classification steps. Pre-processing of the digital slides involves removal of noise and enhancement of the images for better feature highlights. This may include noise smoothening, thresholding, intensity normalization, stain normalization, color separation, etc. After pre-processing, some algorithms may perform a nuclei segmentation task. One of the pre-requisites for breast cancer grading in histopathological images is often the extraction of histopathological anatomical structures like lymphocytes, cancer nuclei, stroma, and background. The shape, size, and other morphological features of these structures are often used as measures for grading and assessing the severity of the disease. Segmentation may also be needed for nuclei counting, which can have some significance for certain types of cancers. Also these nuclei segmentation may be needed for nuclear pleomorphism which has got diagnostic importance in cancer grading [17, 63]. Various segmentation techniques like mean shift, watershed, Gaussian Model, and Active Contour Model [7, 11, 42, 67] are often used for nuclei segmentation.
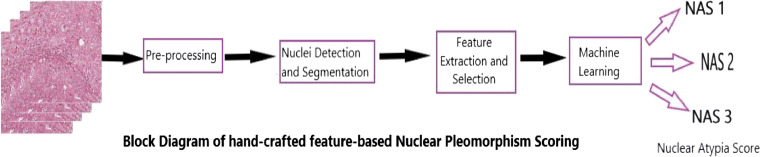
After accurate segmentation, required features capable of distinguishing cancerous and non-cancerous cells are extracted in the feature extraction phase. These features may include the morphological, textural, fractal, or intensity-based features, like the size, shape, number of nuclei, and textural features like histogram, Local Binary Patterns (LBP), and Gray Level Co-occurrence Matrix (GLCM). The extracted features are fed to the classification phase which performs a statistical analysis of these features and often machine learning algorithms are used for segregating these features to different classes. The commonly used classifiers include SVM, Bayesian classifier, k-means clustering, and Artificial Neural Networks(ANN). The block diagram of the steps involved in handcrafted feature-based nuclear pleomorphism is shown in Fig. 5.
Fig. 5.
Block diagram of handcrafted feature-based nuclear pleomorphism scoring
The second category of histopathological cancer grading algorithms mainly involves the learned feature-based algorithms in which high-level feature abstractions are directly extracted from the histopathological images without the need of explicit feature extraction steps. This learned feature-based algorithms have achieved much attention with the success of deep neural networks in various computer vision tasks. These methods are data-driven approaches and hence can be directly transferred for cancer grading and they often outperform the conventional handcrafted approaches. Most of the recently developed algorithms for breast cancer grading belongs to these deep learning–based techniques using Convolutional Neural Networks (CNN), Deep Neural Networks (DNN), Residual Networks (RN), etc.
Compared with CAD applications for radiological images, only a few work has been reported in the quantitative analysis of breast histopathologic grading of stained WSI tissue images. The new era of computer-aided automated breast cancer grading began in 1995 when different morphological features are extracted from the segmented nuclei and used inductive machine learning techniques for atypia scoring by Wolberg et al. [73]. Nuclear features were procured from the manually segmented nuclei and inductive machine learning technique was used for the grading. Most of the works on breast cancer grading were focused on nuclei segmentation for feature extraction as the score is highly related to the morphological aspects of the nuclei in the specimen. This section gives a brief account of the various algorithms used for histopathological breast cancer grading.
Handcrafted Feature-Based Algorithms
Image Pre-processing
Pre-processing suppresses the unwanted distortions in the images and enhances the features that are important for further analysis, like feature extraction. In the image pre-processing step, the artifacts observed in the images may be rectified prior to feature extraction and image analysis. Most of these artifacts are due to the inconsistencies in the preparation of histology slides. Different methods are used in these histological slides to overcome many of the known inconsistencies in the staining process, to bring them into a common, normalized space to enable improved grading of the breast cancer.
In [10], an adaptive approach for color variation removal has been proposed. The pink component that represents the eosin stain of the image is averaged to C vector and the rest of the image is averaged to M vector. Then the H and E components are obtained by the orthogonal projection of M and C, respectively, as given by the following equations:
| 1 |
| 2 |
| 3 |
| 4 |
where Pi denotes the CMY color space vector for the pixel i.
Veta et al. [67] and Basavanhally [5] use color unmixing or color deconvolution, a special case of spectral unmixing, for separation of the stains. In this method, the proportion of each of the different stains applied is calculated based on stain-dependent RGB absorption. For this an orthonormal transformation of the RGB values is done, to get an independent information regarding the contribution of each stain [59]. The major disadvantage of this technique is that areas with multiple stains are treated as one color resulting in loss of information. After color separation, the irrelevant structures that may adversely affect the segmentation accuracy are removed using a series of mathematical morphological operations.
Stain normalization and color separation are done in [42] as a pre-processing step before performing the image analysis. For stain normalization [51], prior information of the stain vectors are used for estimation and unequal stain distribution is dealt with a clustering process, a variant of Otsu thresholding. Trust-region optimization is applied for the underlying optimization task. After normalizing the RGB image, the hematoxylin and eosin stained images are separated through a color deconvolution based on the estimated stain vectors. Then the hematoxylin image which mainly highlights the nuclei is further used for analysis.
Wan et al. [69] use a nonlinear mapping-based stain normalization proposed in [39], to correct the image intensity variations due to variability in tissue preparation. In this method, Principal Color Histograms (PCH) is obtained from a set of quantized image histograms for the evaluation of the color descriptors of the stain. Then this descriptor along with RGB intensity are used for supervised classification by the generation of stain-related probability maps. These probability maps are used for applying a nonlinear normalization of each channel.
In [47], the hematoxylin image is separated by the color deconvolution method proposed in [44]. The RGB color images are mapped to optical density space and subjected to SVD decomposition and a plane corresponding to the two vectors of the largest two singular values of the SVD is calculated. All data is projected onto the plane and normalized to obtain the Optimal Stain Vectors, used for the deconvolution.
Salahuddin et al. [60] used a Pattern-based Hyper Conceptual Sampling based on hyper context feature extraction and reduction, for selecting the most important data from the whole set of training data. Unmixing is done to separate the H&E color channels, followed by anisotropic diffusion filtering to enhance the contrast of the histopathological images in [18] .
Nuclei Detection and Segmentation
The detection and segmentation of nuclei are significant steps in cancer detection, prognosis, and diagnosis. Different aspects of nuclei like its size, morphological structure, and mitotic nuclei count are critical for diagnosing the presence of the disease and for interpreting its severity and malignancy levels. The different nuclei segmentation algorithms used as part of breast cancer grading can be mainly classified as (1) threshold-based techniques, (2) boundary-based techniques, and 3) region growing-based techniques.
Threshold-Based Methods
Image thresholding is considered to be a simple, effective, and widely used method for segregating an image into the foreground objects and background scene. Through selecting a suitable threshold, the histopathological image can be converted to binary image, which contain all of the essential information regarding the shape and size of the nuclear region. This reduces the complexity of the image and simplifies the process of feature extraction and classification. Many of the breast cancer grading algorithms apply this thresholding approach for nuclei segmentation.
Weyn et al. [72] used a combination of background correction and basic thresholding depending on the median of the intensity histogram for extracting the contour of the nuclei resulting in a binary mask, which is embedded into the actual histopathological image for further processing.
A hybrid method of optimal adaptive thresholding along with local morphological operations is used in [53]. A histogram partitioning is used for determining the optimal threshold that will maximize the variance between the classes and minimize the variances within the classes. Grade 3 nuclear segmentation is improved using a further sequence of morphological opening and closing operations combined with the micro structures’ prior knowledge. A similar segmentation technique, that utilizes the adaptive thresholding techniques for the optimal threshold value and standard edge smoothing and morphological filling algorithms, is used in [54].
Naik et al. [49] used a composition of low-level, high-level, and domain-specific information for nuclei segmentation. The Bayesian classifier is applied to generate a pixel-wise likelihood image based on the low-level image intensity and textural information from the RGB image. The intensity values in the likelihood image represents the probability of a pixel belonging to a particular group. The likelihood image is then thresholded to obtain a binary image and the structural constraints are imposed using the knowledge of the domain with respect to the arrangement of histopathological structures. A level set algorithm and template-matching scheme are then used for nuclei segmentation.
In [47], the nuclear region is segmented based on Maximally Stable Extreme Regions (MSER) applied on the hematoxylin contribution map. Multiple thresholds are applied to the image and those areas that change only very little are identified as Maximally Stable Extreme Regions. Then two morphological operations are performed on these regions—opening operation which removes small structures and closing which fills up small holes and breaks in the image.
Boundary-Based Methods
Edges and discontinuity in an image intensity are important characteristics of an image that carry information about object boundaries. Detection methods based on these discontinuities are often used for image segmentation and object identification. Variants of edge-based or boundary-based segmentation methods have been used for nuclear segmentation of breast histopathological images.
In [10], Difference of Gaussian (DoG) filter is applied over the H image, with the size of the filter matching the size of the nuclei. Then Hough Transform is applied to extract the edge map corresponding to various diameters and angles. An Active Contour model is then used for outlining the nuclear boundary. A collection of features that represent the texture, shape, and fitness of the outline are extracted and used for training an SVM classifier.
[12] used morphological operations and distance transform to select the candidate cell nuclei. For this, the gamma-corrected R image is thresholded into a binary image which is subjected to dilation and erosion morphological operations. Distance transform calculated on these images are used for calculating the size of the nuclei. Larger nuclei are avoided as they form part of a clustered tissue and the candidate nuclei are selected for segmentation. Boundary of the nuclei is extracted using polynomial curve fitting on the gradient image obtained from the image patch in the polar space. Then the size and shape features extracted are used for fitting a Gaussian model.
In [32], the nuclear edges are extracted using a snake-based algorithm applied on an image pre-processed with a sequence of thresholding and morphological filtering procedures. ROIs of histological images that include nuclear structures are extracted and then a polar space transformation is performed. An iterative snake algorithm outlines the nuclei boundary on this polar space.
Basavanhally et al. [5] proposed a Color Gradient–based Geodesic Active Contour (CGAC) approach for nuclei segmentation. The optimal segmentation is performed by minimizing an energy function which contains a third term in addition to the two terms in traditional geodesic active contour. This removes the reinitialization phase required for extracting a stable curve that is adopted in traditional methods. The edge detection is performed on the gradient of the gray scale image. The gradients evaluated from each image channel is locally summed up, to obtain the extreme rate of directional changes of the edges. Nuclear regions are extracted out from the boundaries obtained from the CGAC model.
Wan et al. [69] used a combination of boundary and region information, a hybrid active contour method for the automated segmentation of the nuclear region. The hybrid active contour method performs segmentation by reducing the energy function defined as
| 5 |
where represents the image which is to be segmented, denotes the Heaviside function, ω is the image domain, represents the gradient of the image, and α and β are pre-defined weights for the balancing of the two terms. Further, overlapping nuclei are segmented using some local image information. Both local and global image data are used in hybrid active contour model for the better segmentation of nuclear area in digital histopathological slides.
Faridi et al. [18] detected the center of nuclei using morphological operations and DOG filtering and applied the Distance Regularized Level Set Evolution (DRLSE) algorithm for extracting the nuclear boundaries.
Region Growing Methods
Region growing is another methodology used for image segmentation which examines the neighboring pixels based on homogeneity or similarity criteria and added to a region class if the class similarity criteria is satisfied. This process is repeated for each of the pixels surrounding the region. The region growing algorithm starts by selecting a set of seed points based on some criterion. The growing of these regions are then performed from these detected seed points based on some criterion related to region membership. The criterion can be gray level texture, raw pixel intensity, histogram properties, color, etc. Usually, the edges of the regions extracted by region growing are fully connected and are perfectly thin. Region growing techniques are generally better in noisy images where boundaries or edges are difficult to detect. Dalle, Veta, Lu, and Maqlin [11, 42, 46, 67] have used variants of region growing approach for nuclei segmentation for cancer grading.
A multi-resolution approach is used for segmentation in [11]. First neoplasm localization is conducted on the global image with low-resolution. Then cell segmentations are performed on the high-resolution images containing the neoplasmic structures by applying the Gaussian color models. The differences between the Gaussian color distributions are used for detecting the cell types.
A marker-controlled watershed segmentation method is used in [67]. The pre-processed images are used to detect the candidate nuclei, which contains highlighted points having high radial symmetry and regional minima. Watershed segmentation is done starting from these markers and the contour of the nuclei is approximated with ellipses.
In [42], seed detection, local thresholding, and morphological operations are used for the nuclei segmentation. The normalized hematoxylin stain image is then transformed into a blue-ratio (BR) image for easy detection of the nuclear region. A multiscale Laplacian of Gaussian (LoG) performed on this BR images detect the seed points in the image. The scale-normalized LoG-filtered image is fed to a mean-shift algorithm for segmenting the nuclear regions, followed by morphological operations for smoothening the boundaries.
In [46], the peripheral borders of the nuclei are segmented based on a convex grouping algorithm, mainly suitable for open vesicular and patchy type of nuclei which are quite commonly observed in high-risk breast cancers. A k-means clustering algorithm is used for segmenting the nuclear regions which may include these irregular nuclear structures. Then a convex boundary grouping is done to recover the missing edge boundaries.
Plenty of algorithms have been studied and investigated for automated nuclei segmentation. Detecting, segmenting, and classifying the nuclear regions in histopathological images are considered a challenging CAD problem due to the image acquisition artifacts and the heterogeneous nature of the nuclei. Variations in nuclei shape and size and incompleteness in the structure of nuclei also makes the task challenging. The success of the nuclear atypia scoring is highly contingent on the success of the image segmentation technique used, and hence development of a robust algorithm overcoming these issues in a powerful manner, in order to achieve a high level of segmentation accuracy still requires much research to be done.
Feature Extraction and Selection
Disorders in the cell life cycle results in excess cell proliferation in cancer tissues, which result in poor cellular differentiation. It is relevant to obtain various clinically significant and biologically interpretable features from the histopathological images that can better represent the cellular differences in the various grades of cancer. Also, the features should be capable for providing distinguishing quantitative measures for automatic diagnosis and grading of the cancer. Most of these features include morphological, textural, and graph-based topological features. Often, large sets of features are extracted in the hope that a subset of features may include the aspects utilized by human experts for the grading of tumor. And hence, many of the times, the features identified could be irrelevant or redundant. In such cases, a feature selection phase, to select the important and relevant features from this large collection of features, is performed before classification. This section deals with the different features, various feature extraction, and selection methods used in breast histological cancer grading.
Morphological Features
Mainly, the extraction of morphological features depends on the accuracy and efficiency of the underlying segmentation method used. A cancerous cell or nuclei often differ in size and shape when compared with normal one and this difference is made use of by pathologists for cancer grading. The morphological features often deliver details regarding the shape and size of a cell. The size of the segmented nuclei is generally expressed using the radii, perimeter, and area of the nuclei, whereas, the shape is represented by the quantities like smoothness, compactness, symmetry, length of the major and minor axes, roundness, and concavity. Many of the breast caner scoring algorithms have extracted and used these morphological features of the nuclei structures for cancer identification and grading.
Veta et al. [67] used the mean and standard deviation of the nuclear area, extracted from segmented nuclei, for cancer grading and prognosis. A multivariate survival analysis based on Cox’s proportional hazards model revealed that mean value of the nuclear area presented prominent prognostic value whereas the standard deviation of the nuclear region was found not to be an important predictor of the disease outcome.
Petushi et al. [53] extracted the minimum intensity value, area, intensity mean, major and minor axis, standard deviation, and minimum intensity values for each of the segmented nuclei. These 7-features vector is fed for clustering using a pretrained binary decision tree. Various features like area, mean intensity, and circularity of the nuclei and the unfolded nucleoli count are extracted from the boundary segmented nuclear regions in [18].
Different from other scoring algorithms, that mainly concentrate on nuclear pleomorphism for cancer grading, [11] combines all the three criteria in the Nottingham scoring system along with a multi-resolution approach for tumor grading. Tubule formation scoring is performed using the ratio of the total occupied area of the tubule and the total area of the tissue specimen in the histopathological image. For nuclear pleomorphism scoring, the probability distributions of the color values are modeled using the Gaussian functions in all the three grades of cancer. Cell with color distribution closest to the probability distribution of each class is classified of that particular grade. For mitos cell detection, solidity, area, eccentricity, mean, and standard deviation of the intensity values are used for feature vector construction. Gaussian models are constructed for the mitotic and the non-mitotic cell pleomorphism. If the probability of a cell being mitotic is C0 (weighting factor) times greater than the probability of being non-mitotic, then that cell is classified as a mitotic one. Score for mitotic count is calculated depending on the mean of the mitotic count obtained over all the image frames and multiplied by a factor of 10. The overall grade of the cancer is determined based on the scores obtained from tubule formation, nuclear pleomorphism, and mitotic cell count.
Textural Features
Textural features provide important information regarding the variation in intensity of pixel values over a surface in terms of quantities like smoothness, coarseness, and regularity. Textural features are usually extracted using statistical, spectral, and structural methods. The following are the different textural features extracted for histopathologic image analysis and cancer grading.
In [54], micro-texture parameters, which indicate the density of cell nuclei showing dispersed chromatin and the density of cross sections having tubular structure, are identified as potential predictors for the histologic grade of the tumor. These two discriminant features are used for supervised classification techniques like linear classifiers, decision trees, quadratic classifiers, and neural network, of which the quadratic classifiers are shown to have a minimum classification error.
Khan et al. [40] proposed a textural-based feature such as geodesic mean of region covariance descriptors for nuclear grading of breast tumor. They computed region covariance (RC) descriptors for different regions in an image and a single descriptor for the entire image is obtained by deriving the geodesic geometric mean of these RC, known as the gmRC, following a Riemannian trust-region solver-based optimization. The gmRC matrix thus obtained is used for classification based on NGS, using a Geodesic k-Nearest Neighbor classifier(GkNN).
Ojansivu et al. [52] proposed an algorithm based on the textural features for automated classification of breast cancer. They adopted Local Phase Quantization (LPQ) and Local Binary Patterns (LBP) as descriptors, which are used for forming the histograms that represents the statistical textural attributes of the image. The slides are classified into the three grades of cancer using an SVM classifier with a Radial Basis Function (RBF) kernel along with the chi-square distance metric.
In [47], a Bag of Features (BoF) using multiscale descriptors are utilized for representing the detected nuclei. The extracted descriptors are partitioned with the k-means clustering algorithm and used as the atoms of the dictionary. A feature vector obtained from the histogram of these descriptors is used to train an SVM classifier for nuclear pleomorphism grading.
Weyn and Rezaeilouyeh [56, 72] have performed multi-resolution analysis for cancer tissue classification using transform-based textural features like shearlets and wavelets.
The wavelet-based textural features, obtained by repeated low-/high-pass filtering is used in [72], for the representation of chromatin texture in the scoring of malignant breast cancer. From the multiscale representation of the wavelet coefficients, the wavelet-texture features are described as the energies of the image. The classification performance comparison revealed that the wavelet-texture features can perform well comparable with densitometric- and co-occurrence-based features, when used in an automated k-Nearest Neighbor(kNN) classifier.
Rezaeilouyeh et al. [56] computed the shearlet transform on histopathological images and extracted the magnitude and phase of the shearlet coefficients. These shearlet features together with the RGB histopathological image is used to train a CNN with several convolution, max-pooling, and fully connected layers.
Often the morphological and textural features are combined to form a hybrid technique for cancer grading, such as the works of [10, 12, 22, 32, 42, 46].
Cosatto et al. [10] segmented the nuclei outlines and extracted a variety of features that include structural, textural, and fitness of the boundary of the underlying image. Structural features consist of area, symmetry, smoothness, and compactness of edges. Textural features comprises count of vacuoles, variance of H and E channels, and count of nucleoli and DNA strands. They outlined the marked nuclei and discarded malformed ones. Also, the median of the nuclear area in a particular region and the count of the large well-differentiated nuclei in that region are calculated. An SVM classifier is trained based on these extracted features.
Dalle et al. [12] performed nuclear grading based on the shape, size, and texture of the segmented cell nuclei. The size criteria consists of the mean and standard deviation of the segmented cell nuclei. Shape feature includes the roundness of the nuclei and the texture feature is obtained as the mean of the intensity value for each segmented cell nuclei. These features are used as parameters for building a Gaussian model.
Huang et al. [32] use an application-driven image analysis algorithm for high-resolution images and the generic algorithms for the low-resolution images for the analysis, implemented in a multiscale structure supported by sparse coding and dynamic sampling for grading of breast biopsy slides. As a preliminary phase for breast cancer scoring, the most important invasive areas are identified in a low-magnification analysis so as to make the grading process faster. The first- and second-order visual features extracted are used to train a GPU-based SVM, to differentiate between infected areas and the normal tissues. For nuclear pleomorphism assessment, the nuclei are segmented out using iterative snake algorithm from the ROIs identified in the low-resolution analysis phase. A Gaussian distribution is learned from the geometric and radiometric features like size, roundness, and texture extracted from these segmented nuclei, and the scoring is performed using a Bayesian classifier. A multiscale dynamic sampling identifies nuclei with higher pleomorphism, thus avoiding an exhaustive analysis and thereby reducing the computation time.
Lu et al. [42] extracted a group of around 142 morphological and textural features for nuclear grading. These features include nuclear size, mean and standard deviation of stain, sum, entropy, and mean of gradient magnitude image, 3 Tamura texture features, 44 Gray-level run-length matrix-based textural features, and 88 co-occurrence matrix-based Haralick texture features. Each slide is represented using the histogram of each of these features and given to a SVM classifier for grading.
Features that represent the extent of shape and size variations of mitotic nuclei from normal nuclei are used in [46]. The mean and standard deviation of 10 parameters extracted from the segmented nuclei is used as the feature set. These parameters include area, solidity, eccentricity, equivdiameter, average gray value, average contrast, smoothness of the region, skewness, uniformity measure, and entropy.
Gandomkar et al. [22] used a hybrid segmentation-based and texture-based method for extracting features from the histopathological slides that can discriminate the different cancer grades. These cytological features are then combined using an ensemble of trees for regression with the pathologists’ assessment to determine the atypia score of the breast tissues.
Graph-Based Topological Features
Topological or architectural features give information regarding the structure and spatial arrangement of nuclei in a tumor tissue. For that, the spatial interdependency of the cells are represented using various types of graphs, from which the relevant features are extracted for classification. Often, graph-based features are used in combination with morphological or textural features for cancer grading.
Various morphological and graph-based nuclear features, based on the factors used by pathologists for automated grading of breast cancer are extracted out in [49]. The 16 morphological features that represent the shape and size of the segmented nuclei, including the 8 boundary features obtained from the nuclear and lumen structures, are calculated. Also 51 graph-based attributes extracted from the graphical representations like minimum spanning tree, Voronoi diagrams, and Delaunay triangulation are calculated for representing the spatial relationships of the nuclei. Principal Component Analysis (PCA) is done on the features for feature reduction and the high-grade vs. low-grade classification is done using SVM classifier.
Doyle et al. [14] uses a handful of textural- and graph-based features for cancer grading. The textural features include average, minimum-to-maximum ratio, standard deviation, and mode calculated from the second-order co-occurrence Haralick texture features, gray level features, and Gabor filter attributes. The architecture of shape and organization of nuclei within the digital histological slide is represented using graphs like Delaunay Triangulation, Voronoi Diagram, Nuclear density, and Minimum Spanning Trees. The dimensionality of the feature set is reduced by Spectral Clustering (SC) algorithms and fed to an SVM classifier for nuclear scoring.
Basavanhally et al. [5] uses a hybrid set of quantitative features, in which nuclear architecture is represented as graph-based features and nuclear texture as Haralick co-occurrence features. Nuclear architecture is represented using three graphs: Minimum spanning tree, Delaunay triangulation, and Voronoi diagram and features describing variations in these graphs are extracted. Nuclear texture vector is described as the mean, disorder statistics, and standard deviation of the Haralick co-occurrence features. Then the feature selection is obtained via Minimum Redundancy Maximum Relevance (mRMR) for dimensionality reduction and fed to a pretrained random forest for classification.
Wan et al. [69] extract multi-level features for pixel-, object-, and semantic-level information from the breast tumor histopathological images. These pixel-based features include textural features (Kirsch filters, first-order features, Gabor filters, and Haralick features), HoG, and LBP. The object-based features encapsulate the spatial interdependency of nuclei and are represented using Voronoi Diagram (VD), Minimum Spanning Tree (MST), and Delaunay Triangulation (DT). Semantic-level features capture heterogeneity of cancer biology using Convolutional Neural Networks (CNN)–derived descriptors. Dimension reduction is performed using graph embedding and fed to a cascaded ensemble of SVM-based classifiers.
Image Classification
The features extracted from the tumor tissue is a prerequisite for classification or grading of cancer. The classifiers make use of the attributes that represent the nuclear structure and their spatial interdependencies for performing the analysis. Usually classifiers work in two steps: the learning phase and the testing phase. In the learning phase, the features extracted from digital slides with annotation are used for training the classifier. Then these classifiers are tested with unseen data. In the case of nuclear atypia scoring, mainly machine learning algorithms are used to differentiate between the different grades of breast tumors, which includes k-Nearest Neighborhood (kNN) algorithm, decision trees, Support Vector Machines (SVM), Bayes classifiers, Gaussian Mixture models, random forest, and supervised learning techniques. The details regarding the classifiers used for grading of histopathology breast images have been summarized in Table 2. Multi-classifier systems or learning ensembles aggregate predictions of several similar classifiers’ for improving the classification accuracy. Such a cascaded ensemble of three stages of SVM classifiers is used in [69] for classifying the histopathological images into three breast cancer grades. A summary of different handcrafted feature-based algorithms used in literature for Breast Cancer Grading is given in Table 3.
Table 2.
Summary of machine learning algorithms for breast cancer grading
Table 3.
Summary of handcrafted feature-based algorithms used for breast cancer grading
| Paper reference | Pre-processing | Segmentation | Feature extraction | Feature selection | Classification |
|---|---|---|---|---|---|
| Weyn et al. [72] | — | Background correction and basic thresholding | Wavelet-texture features | — | k-nearest neighbor |
| Petushi et al. [53] | —— | Adaptive thresholding followed by morphological operations | Morphological and textural features | Decision tree | |
| Petushi et al. [54] | — | — | Micro-texture parameters | — | Supervised learning |
| Cosatto et al. [10] | Adaptive approach for color variation removal | Hough transform and active contour | Shape, texture, and fitness of the outline | — | SVM |
| Doyle et al. [14] | — | — | Texture- and graph-based features | Spectral clustering | SVM |
| Naik et al. [49] | — | A composition of low-level, high-level, and domain-specific information | Morphological and graph-based features | PCA | SVM |
| Dalle et al. [11] | — | A multi-resolution approach depending on Gaussian color models | Morphological features for the 3 attributes of cancer grading | — | Scoring and grading |
| Dalle et al. [12] | — | Intensity thresholding and line fitting | Size, shape, and textural features | — | Gaussian Mixture Model |
| Huang et al. [32] | — | Iterative snake algorithm | Size, roundness, and textural features | — | Bayesian classifier |
| Veta et al. [67] | Spectral unmixing and morphological operations | Marker controlled watershed segmentation | Mean and standard deviation of nuclear area | — | Cox’s proportional hazards model |
| Basavanhally et al. [5] | Stain-specific RGB absorption and morphological operations | Geodesic active contour | Graph-based and textural features | Minimum redundancy maximum relevance (mRMR) | Random forest |
| Ojansivu et al. [52] | — | — | Local Phase Quantization (LPQ) and Local Binary Patterns(LBP) | — | SVM |
| Lu et al. [42] | Stain normalization and color deconvolution | Mean-shift algorithm and morphological operations | Morphological and statistical texture features | — | SVM |
| Khan et al. [40] | — | — | Geodesic mean of region covariance descriptors | k-nearest neighbor | |
| Maqlin et al. [46] | — | k-means clustering and convex grouping technique | Mean and standard deviation of morphological and textural features | — | Artificial Neural Network |
| Moncayo et al. [47] | Color deconvolution by a linear generative model | Maximally Stable Extreme Regions (MSER) | Multiscale descriptor representing the texture used as Bag of Features (BoF) | — | SVM |
| Faridi et al. [18] | Unmixing of color channels and anisotropic diffusion filtering | DoG filtering and Distance Regularized Level Set Evolution (DRLSE) algorithm | Morphological features and unfolded nucleoli count | — | SVM |
| Wan et al. [69] | Nonlinear mapping-based stain normalization | Hybrid active contour | Textural, graph-based, and CNN-derived features | Graph embedding | SVM-based cascaded ensemble of classifiers |
| Salahuddin et al. [60] | Pattern-based Hyper Conceptual Sampling | — | Hyper context feature extraction | — | SVM, Naive Bayes, Feed Forward Net (FFN), Cascade Forward Net (CFN), and Pattern Net(PN) |
| Gandomkar et al. [22] | Stain normalization and color deconvolution | Morphological operations followed by thresholding | Textural features like first-order statistics, Haralick, LBP, gray-level run-length matrix, maximum response filter, and Gabor-based features | — | Multiple regression trees |
Automated handcrafted feature-based breast cancer grading or prognosis system relies on the disease-related features extracted from the histopathological images. This may require accurate detection and segmentation of nuclear structures, which itself is a challenging task because of the complexity and high density of histologic data. Consequently, there is a great demand for computational and intelligent systems for nuclear pleomorphism analysis. Recently developed deep learning (DL) techniques can extract and organize discriminative information about the data thereby avoiding the need of these handcrafted features. Many of the deep learning systems like Convolutional Neural Networks (CNNs) are found to be successful in various classification tasks such as object recognition, signal processing, speech recognition, and natural language processing.
Learned Feature-Based Algorithms
Various works like [3, 4, 29, 50, 55] have investigated applying Convolutional Neural Networks for nuclear atypia scoring and were found to perform better than systems that use handcrafted feature descriptors. Han et al. [29] propose an exhaustive recognition technique with a class structure-based deep convolutional neural network (CSDCNN) for multi-classification of the histopathological breast cancer images. The CSDCNN learns discriminative and semantic hierarchical features and the feature similarities of different classes are specified using feature space distance constraints assimilated into the network model. Rakhlin et al. [55] use a Gradient Boosting algorithm on top of the pretrained CNN on ImageNet for the classification of H&E stained breast cancer images. Bardou et al. [4] extract the handcrafted features (bag of words and locality constrained linear coding) and classified the cancer subtypes using CNN with a fully connected classifier layer and [50] uses a single-layer Convolutional Neural Network (CNN) for the binary classification of the microscopic breast cancer images. Araujo et al. [3] used CNN architecture designed to extract details at various scales in the nuclei and tissue level.
Wollmann et al. [6, 8, 26, 48, 56, 61, 62, 69, 75, 76] used deep learning technique-based CNN architectures for the classification of breast histopathological images. Wollmann et al. [75] performs slide level classification using deep neural networks on the patches cropped by color thresholding and these results are aggregated to grade the patient. Spanhol et al. [61] used an existing CNN architecture, AlexNet, modified for the grading problem in breast cancer. The image patches extracted from high-resolution BreaKHis dataset are used for training the deep networks and an integration of these patches is adopted for the final classification. The image patches are extracted mainly based on two strategies—using a sliding window allowing 50% of overlapping and a random choice of non-overlapped patches. Different networks are trained with different number of patches having various sizes. Hence, the results are improved through a combination of classifiers. The final layer in AlexNet is the fully connected layer using softmax activation which estimates the output as posterior probabilities. Different combination or fusion rules are applied for combining the posterior probability estimations from different classifiers using Sum, Product and Max rules. The fusion using the Max rule is found to outperform the Sum and Product rules.
Wan et al. [69] used CNN-derived semantic-level descriptors together with pixel-based and object-based features for atypia scoring. They proposed a 3-layer CNN model comprising of two successive convolutional and max-pooling layers and a fully connected classification layer. The two consecutive convolutional and the max-pooling layers adopt the same fixed 9× 9 convolutional kernel and 2×2 pooling kernel. The final layer has 38 neurons, which are connected to the last three neurons corresponding to three classes—low-grade, intermediate-grade and high-grade cancer. The semantic-based features are generated using this CNN trained with labeled and segmented nuclei of various grades.
Rezaeilouyeh et al. [56] used the shearlet coefficients, especially the magnitude and phase, as subsidiary information for the neural network along with the original image for the CNN-induced cancer classification. Shearlet transform is a multiscale directional representation exhibiting affine properties and can extract anisotropic features at various scales and orientations. The shearlet transform coefficient contains a magnitude and a phase part. Phase part contains most of the information and is invariant to the induced noise and to the variations in the image contrast. Magnitude part of the shearlet coefficients mainly represents the singularities or edges in the image. Since breast histopathological images are stained with H&E, the RGB color data together with the magnitude and phase of the shearlet coefficients are used for training the CNNs. The magnitude and phase of shearlets from different decomposition levels and the RGB images are fed to separate CNNs as they have different properties. The proposed CNN consists of three layers of convolution and pooling, with the convolutional layer using 64 Gaussian filters with a size of 5×5 having a standard deviation of about 0.0001 and a bias of zero and the max-pooling layer applied on 3×3 region with a step size of 2 pixels. The Rectified Linear Unit (ReLU) is adopted as the activation function and a fully connected layer is used for combining the outputs from different CNNs fed with various features.
Xu et al. [76] proposed a Multi-Resolution Convolutional Network (MR-CN) with Plurality Voting (MR-CN-PV) model for automated nuclear atypia scoring. The model consists of a combination of three Single-Resolution Convolutional Network based on AlexNet, each performing independent scoring based on majority voting on three different resolutions of images. Score from these three SR-CN-MV are then integrated with plurality voting strategy for the final atypia scoring.
Bayramoglu et al. [6] proposed a multi-task CNN architecture that can predict both the image magnification level and its malignancy property simultaneously. The proposed model allows combining image data from many more resolution levels than four discrete magnification levels. The proposed CNN consists of three separate convolutional layers, with each one followed by the ReLU operation and a max-pooling layer. After the convolution layers, two fully connected layers are added. For multi-tasking, the network is modified by splitting the last fully connected layer into two branches. The first branch, which classifies tissues based on malignancy, is fed to the 2-way softmax, at the same time, the second branch, which learns the magnification factor, is fed to a 4-way softmax layer and the softmax loss is minimized.
Nahid et al. [48] classify the breast cancer images using three deep neural network models: CNN, a Long-Short-Term-Memory (LSTM), and an integrated CNN+LSTM model, guided by an unsupervised clustering that extract out the structural and statistical information from the histopathological images. The extracted global and local features are used for deciding the class by the Softmax and SVM layers. Bejnordi et al. [8] use a stack of context-aware CNNs to extract the information regarding the interdependence of cellular structures in the WSIs of benign, DCIS, and IDC. Guo et al. [26] propose a hybrid CNN architecture with two CNN models: a patch-level voting module and a merging module and uses hierarchy voting tactic and bagging technique for reducing generalization error. The data augmentation and transfer learning are used to increase the number of training data.
The results on using deep neural networks for cancer grading show that they can give higher classification accuracy than handcrafted features, but that may require complex systems with long training times and highly efficient fine tuning mechanisms. An in-between solution to handcrafted and task-specific CNN methods, the DeCAF(A Deep Convolutional Activation Feature) features, is proposed in [62] for breast cancer classification task. It gives the advantage of reusing a pretrained CNN for feature extraction, and these features are used as input for a new classifier trained on a problem specific data.
Other variants of CNN architectures like deep belief networks [46], BiCNN model [70], residual networks [2, 21, 66] and Inception networks [2, 65, 66] are also used for breast cancer grading. Maqlin et al. [46] proposed a deep belief network (DBN-DNN) for the breast cancer grading. Each layer of the DBN-DNN structure consists of Restricted Boltzmann Machines (RBM). RBMs are artificial neural networks which are generative, and they learn the probability distribution from the input set. The RBMs are trained through a Contra Divergence (CD) algorithm applied in an unsupervised manner. A sequence of such RBMs are stacked together to derive a DBN, by cascading the states of the hidden nodes of the previous level as the inputs to the next level of RBMs. Then a final layer is added to this DBN to construct a DNN. A classical back-propagation algorithm applied in a supervised way is used for fine-tuning the constructed DBN-DNN. The histological images are segmented and 20 features are extracted out of them. These features are used for training the DBN-DNN classifier having a 20-13-13-3 architecture, with dual layers of RBM each having 13 hidden layers.
Wei et al. [70] proposed a deep convolutional neural network, named as BiCNN model, for breast cancer classification on the pathological image. BiCNN is structured as a series of the stages: input layer, convolution layer, pooling layer, and a softmax classifier with loss. To overcome over-fitting of data, raw BreakHis database is augmented 14 times by rotation, scaling, and mirroring. Two training strategies are employed—BiCNN from scratch and transfer learning in which unsupervised pretraining is performed on ImageNet and fine-tuning on BreakHis using BiCNN.
Gandomkar et al. [21] proposed a DEep Residual Network (MUDeRN) for multi-classification of breast histopathological images. The malignancy of the cancer is determined using a ResNet (Residual Network) and in the next stage, they are classified into corresponding subtypes and eventually the outputs of the ResNets are combined using a meta-decision tree (MDT). Alom et al. [2] combined the potential of the Residual Network (ResNet), the Inception Network (Inception-v4), and the Recurrent CNN (RCNN) to design an Inception Recurrent Residual CNN (IRRCNN) that can provide a better performance for the breast cancer classification than its constituent networks.
Vang et al. [65] used the Inception V3 model for discriminating the tissue patches and the image-level prediction is performed using a two-stage ensemble fusion framework involving Dual Path Network (DPN) for feature extraction and Gradient Boosting Machine (GSM) and logistic regression for the final prediction.
Vesal et al. [66] utilized the robustness of transfer learning for the breast cancer classification. They used the ResNet50 and Google’s Inception-V3 networks pretrained on ImageNet and the pretrained weights are fine-tuned to learn the features pertaining to the histopathological classification task. Golatkar et al. [25] performed the fine-tuning of the Inception-v3 CNN for the histopathological breast cancer classification, using the patches extracted based on nuclear density and majority voting is employed for the final classification of the image. Jannesari et al. [34] used ImageNet Inception and ResNet pretrained models networks for the classification of different types of breast cancer.
Jiang et al. [35] combined Squeeze-and-Excitation block (SEblock) and the residual block of ResNet for the multi-class classification of breast cancer, that reduce number of parameters for the model training and circumvent the model overfitting problem. The learned feature-based algorithms used in literature for Breast Cancer Grading is summarized in Table 4.
Table 4.
Summary of learned feature-based algorithms for breast cancer grading
| Learning algorithm used | Published papers | Description | Advantages | Disadvantages |
|---|---|---|---|---|
| Convolutional Neural Network (CNN) | Han et al. [29] | Class structure-based convolutional neural network (CSDCNN) | Better feature learning capability as feature space distance constraints integrated into CSDCNN | Needs extensive training with augmented data. |
| Nejad et al. [50] | Single-layer CNN | Much faster and less resource demanding | Recognition rate is less | |
| Araújo et al. [3] | CNN extracts nuclei and tissue level features | Retrieve information at different scales | Class imbalances not considered | |
| Rakhlin et al. [55] | Gradient Boosting algorithm on top of the pretrained CNN on ImageNet | No network training as it uses a pretrained model | Requires pre-processing of the images | |
| Bardou et al. [4] | CNN classifies the features extracted by bag of words and locality constrained linear coding | Combines both the handcrafted, feature-based, and learned feature-based algorithms | Lower performance for the multi-class classification | |
| Deep Learning | Wollmann and Rohr [75] | Deep Neural Network | Fast processing speed | Cellular information loss on downsampling |
| Spanhol et al. [61] | Modified AlexNet | Introduces a real life, challenging dataset | High false positive rate | |
| Rezaeilouyeh et al. [56] | DNN fed with Shearlet Coefficients | Accuracy improved using the magnitude and phase of shearlet coefficients | Highly complex | |
| Wan et al. [69] | CNN-derived semantic-level descriptors ‘ | Uses multi-level features | Needs segmenting the nuclei which forms a challenging task | |
| Xu et al. [76] | Multi-Resolution Convolutional Network (MR-CN) with Plurality Voting (MR-CN-PV) | Integrate image features from multiple fields of views (FOV) | Multiple training required for each field of view | |
| Bayramoglu et al. [6] | Multi-task CNN | Classify histopathology images independent of their magnifications | Only patches from image center are used which may fail to assimilate the information of the whole image | |
| Bejnordi et al. [8] | Stack of context-aware CNNs | Incorporate global interdependence information of cellular structures to facilitate predictions | Increased computational cost | |
| Spanhol et al. [62] | DeCAF(A Deep Convolutional Activation Feature) | Pretrained CNN for representation learning | Not suitable for images with more fine-grained structures | |
| Nahid et al. [48] | Integrated CNN and Long-Short-Term-Memory (LSTM) model | DNN model guided by the cell nuclei structural and statistical information | Dataset comparatively smaller for a DNN | |
| Guo et al. [26] | Hybrid CNN with hierarchy voting tactic and bagging technique | Reduced generalization error and improved performance | Low sensitivity | |
| CNN variants | Maqlin et al. [46] | Deep belief network | Faster and accurate | Require handcrafted feature extraction prior to CNN learning |
| Wei et al. [70] | BiCNN model | Good robustness and generalization | Computational burden | |
| Gandomkar et al. [21] | Residual network | Aggregate image-level classification and produce patient-level diagnosis | Multiple training stages, performance degrades at a different magnification level for the test images | |
| Alom et al. [2] | Residual network and Inception network | Magnification factor invariant binary and multi-class classification | Computational cost | |
| Vesal et al. [66] | Residual network and Inception network | Uses transfer learning technique | Requires fine tuning of the network parameters | |
| Vang et al. [65] | Inception network | Image classification based on smaller sized patches | Further processing required to improve the sensitivity | |
| Golatkar et al. [25] | Inception-v3 CNN | Uses pretrained networks | Discrimination capability less | |
| Jannesari et al. [34] | Residual network and Inception network | High sensitivity and negligible false negative and false positives | Requires fine tuning of parameters | |
| Jiang et al. [35] | SE-ResNet | Reduced parameter set | Stacked SE-ResNet makes the network more complex |
Another learning-based paradigm that has attracted much attention in the area of computer vision is the dictionary learning–based techniques. While deep learning techniques learn the features of the data by updating the weights of the layers, the dictionary learning aims in learning the basis features from the data by matrix factorization. A sparse coding and dictionary learning–based approach implemented over the symmetric positive definite (SPD) matrices has been used for breast cancer grading in [13]. The dictionary learning task on the SPD manifold is recast into a high-dimensional Hilbert space and is observed to be superior in discriminating the cancer grades.
Evaluation Metrics
A classification system usually includes two stages: (i) training stage and (ii) testing stage. Training involves determining the parameters and building a model for the classification using a training set of data. After training, the system performance is measured, using a set of unseen data, known as test set, by holding the learned parameters to be constant. As more data is used for training, system will be more better designed, and if more data is used for testing, we can have a more reliable evaluation of the system. Before using the trained network for real-time applications, there is always a need to validate the stability of the system using a dataset not used for training. This is done by cross-validation using a validation dataset which is created from the training set, to fine-tune the model and to make sure that there is no overfitting of the data. For this, the validation set is input to the system and checked if the error is within some range.
Main types of cross-validation techniques involve holdout, k-fold, leave-one-out, and leave-p-out cross-validations. In the case of holdout cross-validation, p*100% of the data is randomly chosen for validation purpose, and the system is trained using the remaining data. In the case of k-fold cross-validation, training data is partitioned randomly to k groups. Then, the k–1 groups are used for the training of the system and the remaining group is used for estimating the error rate. This process is repeated for k number of times so that each of the groups is considered for the testing of the system. In the case of leave-p-out cross-validation, p data points are left out of training data for validation, i.e, out of n data, n-p are used for training, and p of them used for validation. The leave-p-out cross-validation having p = 1 forms the leave-one-out cross-validation; therefore, only a single data is used for estimating the error rate of each step.
As it is required to quantitatively measure the classification accuracy of the system, on unknown samples, the test set should be chosen with care so as to include samples that are often different and independent from those samples adopted for training and validation. For a given test data, there are four possibilities:
True-Positive (TP): positive cases that are correctly identified by the classifier.
True-Negative (TN): negative cases that are correctly identified by the classifier.
False-Positives (FP): negative cases that are incorrectly identified as positive.
False-Negatives (FN): positive cases that are incorrectly identified as negative.
In the case of nuclear atypia scoring, it is a multi-classifier problem with 3 classes, NAS 1, NAS 2, and NAS 3. In such a case, the TP, TN, FP, and FN are calculated for NAS 1 as
“TP of NAS 1” all NAS 1 cases that are classified as NAS 1.
“TN of NAS 1” all non-NAS 1 cases that are not classified as NAS 1.
“FP of NAS 1” all non-NAS 1 cases that are classified as NAS 1.
“N of NAS 1” all NAS 1 cases that are not classified as NAS 1.
TP, TN, FP, and FN for NAS 2 and NAS 3 can be calculated similarly.
The evaluation metrics are used for evaluating the generalization capability of the trained classifier model and also as an evaluator for model selection. They are used to measure and summarize the performance of trained classifier when tested with the unseen test data. Accuracy is one of the most common metrics used by many researchers. It measures the ratio of correct classifications and the total cases of test data evaluated.
| 6 |
Other widely used evaluation metrics [31] include error rate, sensitivity, specificity, precision, negative predictive value (NPV), false positive rate (FPR), false negative rate (FNR), F1-score, and Matthew’s Correlation Coefficient (MCC). Error rate measures the ratio of incorrect classifications over the total number of test data evaluated. Sensitivity or true positive rate (TPR) or recall measures the number of positives that are correctly classified, whereas specificity or true negative rate (TNR) calculates the number of negatives that are correctly classified. Precision or positive predictive value (PPV) is used for measuring the positives which are correctly identified out of the total samples in the positive class. Negative predictive value (NPV) is the portion of negatives which are correctly identified out of the total samples in the negative class. False positive rate (FPR) is measured as the ratio of the number of negative samples wrongly predicted as positive (i.e, false positives) and the total number of actual negative samples. False negative rate (FNR) is measured to be the ratio of the number of positive samples wrongly predicted as negative (i.e, false negatives) and the total number of actual positive samples. F1-score is obtained from the harmonic mean of sensitivity and precision values. Matthew’s Correlation Coefficient (MCC) is calculated as the correlation coefficient of the observed classifications against the predicted classifications, and its value is between − 1 and + 1. A value of + 1 denotes a perfect prediction, whereas 0 denotes a uniform random classification and − 1 denotes an inverse prediction. Higher the accuracy, precision, sensitivity, specificity, NPV, and F1-score, the more successful is the classification system.
| 7 |
| 8 |
| 9 |
| 10 |
| 11 |
| 12 |
| 13 |
| 14 |
| 15 |
Another useful technique for organizing classifiers and visualizing their performance is using Receiver Operating Characteristics (ROC) graphs. An ROC curve depicts the relative trade off between the true positive rate (TPR) against the false positive rate (FPR) for different thresholds. TPR indicates sensitivity and FPR is similar to 1-specificity. The area under the curve (AUC) can be calculated to measure the ability of the extracted features in differentiating the breast cancers of different grades. Greater the area under the curves, the better the system. Figure 6 shows a typical ROC curve for different models. In the case of random classification, a classifier with an AUC value greater than 0.5 is considered better, 0.7 to 0.8 is considered fair, 0.8 to 0.9 is good, and greater than 0.9 is treated as outstanding.
Fig. 6.
ROC curves for different models [37]
Being nuclear pleomorphism scoring, a multi-classification task, it grades breast tumor into 3 classes and hence different ROC graphs need to be plotted, one per each class. So, if C denotes the set of all the 3 classes, ROC graph i depicts the classification performance for class ci with ci considered to be the positive class and with all other classes considered negatives [19], i.e.,
| 16 |
| 17 |
For AUCs of multi-class problems, each class specific ROC curve is drawn; the AUC for each graph is measured and then the AUCs are weighted using the prevalence of the reference class in the data denoted as p(ci) and the obtained values are summed up, i.e,
| 18 |
where AUC(ci) represents the area measured from the class reference ROC curve of ci.
Comparative Analysis of Nuclear Atypia Scoring Algorithms
Histopathological image analysis for cancer grading has been a difficult and complicated task compared with other medical image processing and analysis methods. This may be due to the artifacts present in the histological images and due to the complex structural patterns of cancer tissues. Algorithms have been developed for automated grading of breast cancer that shows remarkably exciting results. A comparative study of the different techniques summarized in the literature is tiresome as each of the methods adopts their own distinctive dataset and the results are presented with divergent evaluation metrics. So, a comparative analysis of some typical algorithms for breast cancer nuclear atypia scoring based on a common dataset applied on these algorithms is discussed in this section.
The algorithms used for the comparative study include the four techniques for breast cancer grading as discussed in [40, 42, 56] and [76]. Lu, Khan, and Rezaeilouyeh [40, 42, 56] and Xu [76] use the morphological features, textural features, transform-based features, and deep learning techniques, respectively, for cancer grading. Of them, the Khan and Lu methods are found to be bestowing the best performance in the handcrafted feature category of algorithms, while the Rezaeilouyeh and the Xu methods in the learned features category and hence they are selected for the comparative study. Among them, the Khan method has achieved the first place in the MITOS-ATYPIA14 challenge in the ICPR2014.
In [42], initially stain normalization and color separation is performed on the H&E images. The separated H image is converted to a blue-ratio image, which is used for nuclear segmentation. Scale-normalized Laplacian of Gaussian (LoG) filtering is performed on this blue-ratio image and the filtered image is processed with a mean-shift algorithm which detects the seed points of the nuclei. Morphological operations are applied on these seed points to obtain the segmented nuclei, from which 142 morphological and textural features are extracted. The histogram of each features is used to represent each image. These features are applied to an SVM classifier for nuclear atypia scoring.
A novel image-level descriptor based on region covariances is presented in [40] for grading of breast whole slide images. The images are subdivided into several non-overlapping regions and region covariances (RC) are computed for each of them. The textural features for calculating the RC are extracted using Maximum Response 8 (MR8) filter banks. These RC descriptors extracted represent symmetric positive definite (SPD) matrices, and hence, they form points embedded on the Riemannian manifold. The region-level RC descriptors are then combined to get a single descriptor at the image-level for the whole image obtained as the geodesic geometric mean of these region covariances, known to be gmRC descriptors. Geodesic geometric mean of the covariance matrices is computed adopting the Frechet mean calculation of the RC descriptors by the optimization through the trust-region solving in the Riemannian manifolds. These gmRC descriptors are used for breast cancer grading using a geodesic k-nearest neighbor (GkNN) classifier. GkNN computes the distances among the SPD matrices on the Riemannian manifold using three distance metrics: Log-Euclidean metric, affine-invariant metric, and Stein divergence.
Rezaeilouyeh et al. [56] proposed the use of phase values of shearlet coefficients as a key feature for breast cancer grading. Shearlet transform [15] is a multiscale directional representation system exhibiting affine properties and can provide the sparse approximation for the anisotropic features at various scales and orientations. Shearlet transform is applied on the histopathological images and then the magnitude and phase of shearlet coefficients are calculated. The singularities or edges of the image can be represented by the magnitude of the shearlet coefficients, while most of the image’s information is contained in the phase. The RGB original image of the digital slide is also used along with the shearlet magnitude and phase, as the details of the color is also important for cancer detection since the tissue slides are H&E stained. Then the most relevant feature representations are directly learned from the images using Convolutional Neural Networks (CNN), which includes different layers of convolutions preceded by pooling layers together with dropout applied on fully connected layers.
Xu et al. [76] present a deep learning–based approach for automated Nuclear Atypia Scoring (NAS) on breast histopathology. A Multi-Resolution Convolutional Network with Plurality Voting (MR-CN-PV) is used for the automated scoring. A digital pathology slide is scanned at different resolution of × 10, × 20, and × 40. The deep learning model consists of a combination of three Single-Resolution Convolutional Network with Majority Voting (SR-CN-MV) paths, which performs NAS through majority voting independently. AlexNet based network is used as the CNN model. Each of the SR-CN-MV path is independently trained with the slides scanned at three resolutions. The grading obtained from the SR-CNN are then combined with plurality voting strategy for obtaining the final score.
The breast cancer histopathological images for the experiments are obtained from the publicly available dataset released as part of the MITOS-ATYPIA14 7 challenge carried out as part of the International Conference on Pattern Recognition (ICPR2014). The dataset released for the nuclear atypia scoring consists of histopathological slides stained using (H&E) dyes and are scanned with the Aperio Scanscope XT and Hamamatsu Nanozoomer 2.0-HT scanners. The images are obtained from 11 different invasive carcinoma infected personnels and from each of these images, ×20 magnified frames are extracted and used for grading by two independent pathologists. Those frames with conflict in score are further annotated by a third expert and the majority score is assigned on a scale of 1 to 3. NAS is performed on both set of images from Aperio and Hamamatsu scanners separately and combined. The above mentioned four algorithms are used for breast cancer grading and their performance is compared based on the evaluation metrics described in the “Evaluation Metrics” section.
The algorithms are implemented on MATLAB and the evaluation metrics are calculated following a five fold cross-validation repeated over 10 times and the average performance values are calculated and reported. The performance comparison results of Atypia on Aperio dataset are summarized in Table 5. As per the results, morphological feature-based algorithm, [42] could not produce satisfactory results, as the algorithm makes use of the segmented nuclei for extracting the nuclear features. Hence, the algorithm heavily relies on the precise segmentation of the nuclei. Segmentation of nuclei in histopathological images itself is a great challenge as often the nuclear boundaries are not well defined and the chances of overlapping nuclei is high in high-grade tumors. However, when compared with linear, polynomial, and sigmoid functions, the best performance with a classification accuracy of 78% for the [42] algorithm was obtained when Radial Basis Function (RBF) was applied on the SVM classifier.
Table 5.
Performance comparison of atypia scoring on Aperio dataset
| Algorithm | Classifier | Accuracy | Error | Sensitivity | Specificity | Precision | NPV | FPR | FNR | F1_score | MCC |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Lu et al. [42] | SVM-Linear | 0.7500 | 0.2500 | 0.3333 | 0.6667 | 0.2500 | 0.5833 | 0.3333 | 0.6667 | 0.2857 | 0.1635 |
| SVM-Poly | 0.7433 | 0.2567 | 0.4339 | 0.7570 | 0.4047 | 0.7880 | 0.2430 | 0.5661 | 0.4186 | 0.2036 | |
| SVM-RBF | 0.7800 | 0.2200 | 0.4551 | 0.7718 | 0.4455 | 0.8348 | 0.2282 | 0.5449 | 0.4476 | 0.2628 | |
| SVM-Sigmoid | 0.7500 | 0.2500 | 0.3333 | 0.6667 | 0.2005 | 0.5833 | 0.3333 | 0.6667 | 0.2857 | 0.1635 | |
| Khan et al. [40] | GkNN- logE | 0.8347 | 0.1653 | 0.6755 | 0.8315 | 0.8038 | 0.8711 | 0.1685 | 0.3245 | 0.6975 | 0.5692 |
| GkNN-affine | 0.8293 | 0.1707 | 0.7019 | 0.8342 | 0.7851 | 0.8663 | 0.1658 | 0.2981 | 0.7197 | 0.5833 | |
| GkNN-Stein | 0.8213 | 0.1787 | 0.6695 | 0.8261 | 0.7784 | 0.8601 | 0.1739 | 0.3305 | 0.6992 | 0.5562 | |
| Rezaeilouyeh et al. [56] | Neural Network | 0.7500 | 0.2500 | 0.3333 | 0.6667 | 0.2500 | 0.5833 | 0.3333 | 0.6667 | 0.2857 | 0.1635 |
| Xu et al. [76] | Deep learning | 0.8000 | 0.2000 | 0.6646 | 0.8156 | 0.7352 | 0.8461 | 0.1844 | 0.3354 | 0.6879 | 0.5264 |
The shearlet transform-based algorithm discussed in [56] also gives similar results with a classification accuracy of 75%. Here, the magnitude and phase of shearlet transform-based features, which are image-level descriptors, fail to represent the heterogeneity which can be observed to be distributed over the histopathological images. The deep neural network–based algorithm [76] gives a better and comparable result for nuclear atypia scoring with a classification accuracy of 80.0%. It is observed that the deep learning techniques can achieve much better results than the use of handcrafted morphological and textural feature descriptors, which indicates that these deep learning techniques are a viable alternative for a breast cancer grading systems. The results show their ability to discover and learn nuclear features from raw histopathological images through feature learning. The advancements in computing power and availability of large benchmarked datasets can unleash and exploit the capabilities of neural networks through much deeper architectures.
The results of NA scoring on Hamamatsu dataset are summarized in Table 6. The algorithm using geodesic geometric mean of the region covariance [40] gives the best results compared with the others with a maximum classification accuracy of 83.5% and 82.1% for Aperio and Hamamatsu scanners, respectively. The low-dimensional RC descriptors extracted at the region level has represented the heterogeneity of the histopathological images effectively, where regions of similar morphological architecture is seldom found. Also the generalized geodesic geometric mean of RCs offers an efficient method for combining multiple covariance descriptors from the same image. Results show that the covariance of a histopathologic image is highly discriminative enough to represent the nuclear pleomorphism. Moreover, their dimensionality is comparatively less, thereby the computational cost is reduced irrespective of the size of the image regions.
Table 6.
Performance comparison of atypia scoring on Hamamatsu dataset
| Algorithm | Classifier | Accuracy | Error | Sensitivity | Specificity | Precision | NPV | FPR | FNR | F1_score | MCC |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Lu et al. [42] | SVM-Linear | 0.7475 | 0.2525 | 0.3333 | 0.6667 | 0.2492 | 0.5825 | 0.3333 | 0.6667 | 0.2852 | 0.1732 |
| SVM-Poly | 0.7643 | 0.2357 | 0.3931 | 0.7049 | 0.6197 | 0.8287 | 0.2951 | 0.6069 | 0.3990 | 0.2095 | |
| SVM-RBF | 0.7576 | 0.2424 | 0.4066 | 0.7263 | 0.4234 | 0.8005 | 0.2737 | 0.5934 | 0.4023 | 0.1817 | |
| SVM-Sigmoid | 0.7475 | 0.2525 | 0.3333 | 0.6667 | 0.2492 | 0.5825 | 0.3333 | 0.6667 | 0.2852 | 0.1732 | |
| Khan et al. [40] | GkNN- logE | 0.8135 | 0.1865 | 0.6212 | 0.8021 | 0.6831 | 0.8434 | 0.1979 | 0.3788 | 0.6229 | 0.4616 |
| GkNN-affine | 0.8189 | 0.1811 | 0.6507 | 0.8178 | 0.7831 | 0.8707 | 0.1822 | 0.3493 | 0.6832 | 0.5460 | |
| GkNN-Stein | 0.8203 | 0.1797 | 0.6703 | 0.8188 | 0.7081 | 0.8527 | 0.1812 | 0.3297 | 0.6608 | 0.5134 | |
| Rezaeilouyeh et al. [56] | Neural Network | 0.7475 | 0.2525 | 0.3333 | 0.6667 | 0.2492 | 0.5825 | 0.3333 | 0.6667 | 0.2852 | 0.1732 |
| Xu et al. [76] | Deep learning | 0.7973 | 0.2027 | 0.6925 | 0.8018 | 0.6899 | 0.8207 | 0.1982 | 0.3075 | 0.6838 | 0.4999 |
Almost all the four algorithms show a consistent result for both the datasets from Aperio and Hamamatsu scanners as shown in Tables 5 and 6. Figures 7 and 8 shows the corresponding bar charts. Further experiments were conducted for these algorithms combining both Aperio and Hamamatsu dataset whose results are given Table 7 and Fig. 9. Here also, the gmRC-based method shows the best classification accuracy of 83.3%. However, these results are not satisfactory enough to be adopted for clinical use, as covariance matrices are prone to be singular and having a fixed form of representation, with limited capability in modeling complicated cancer nuclear features. This indicates that improvements are still required to develop a robust and accurate technique suitable to be adopted for clinical applications. The adoption of an automated cancer grading system for pathological applications will be a milestone in the field of medical diagnosis.
Fig. 7.
Performance comparison of atypia scoring on Aperio dataset
Fig. 8.
Performance comparison of atypia scoring on Hamamatsu dataset
Table 7.
Performance comparison of atypia scoring on combined Aperio and Hamamatsu datasets
| Algorithm | Classifier | Accuracy | Error | Sensitivity | Specificity | Precision | NPV | FPR | FNR | F1_score | MCC |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Lu et al. [42] | SVM-Linear | 0.7487 | 0.2513 | 0.3333 | 0.6667 | 0.2496 | 0.5829 | 0.3333 | 0.6667 | 0.2854 | 0.1789 |
| SVM-Poly | 0.7705 | 0.2295 | 0.4340 | 0.7373 | 0.4572 | 0.8074 | 0.2627 | 0.5660 | 0.4346 | 0.2225 | |
| SVM-RBF | 0.7487 | 0.2513 | 0.3333 | 0.6667 | 0.2496 | 0.5829 | 0.3333 | 0.6667 | 0.2854 | 0.1789 | |
| SVM-Sigmoid | 0.7487 | 0.2513 | 0.3333 | 0.6667 | 0.2496 | 0.5829 | 0.3333 | 0.6667 | 0.2854 | 0.1789 | |
| Khan et al. [40] | GkNN- logE | 0.8329 | 0.1671 | 0.6771 | 0.8326 | 0.7676 | 0.8649 | 0.1674 | 0.3229 | 0.7085 | 0.5649 |
| GkNN-affine | 0.8195 | 0.1805 | 0.6752 | 0.8237 | 0.7464 | 0.8576 | 0.1763 | 0.3248 | 0.6911 | 0.5432 | |
| GkNN-Stein | 0.8208 | 0.1792 | 0.6650 | 0.8245 | 0.7605 | 0.8645 | 0.1755 | 0.3350 | 0.6947 | 0.5495 | |
| Rezaeilouyeh et al. [56] | Neural Network | 0.7487 | 0.2513 | 0.3333 | 0.6667 | 0.2496 | 0.5829 | 0.3333 | 0.6667 | 0.2854 | 0.1789 |
| Xu et al. [76] | Deep learning | 0.7987 | 0.2013 | 0.7649 | 0.8525 | 0.6683 | 0.8242 | 0.1475 | 0.2351 | 0.7065 | 0.5509 |
Fig. 9.
Performance comparison of atypia scoring on combined Aperio and Hamamatsu datasets
Moreover, the existing state-of-the-art algorithms for histopathological grading based on handcrafted features and learned features often exploit the Euclidean geometry of the underlying histopathological image samples. Recent advancements in machine learning and computer vision suggest considering the non-Euclidean geometry of the problem for addressing a wide extent of problems. Hence, by transforming the histopathological images to a non-Euclidean space may procure better solution for the automated nuclear atypia scoring.
Nuclear Atypia Scoring: Future Perspective
Digital imaging in pathology has emerged and gone through an exponential growth catalyzed by the advancements in the field of histopathological imaging techniques, storage capacity, database management mechanisms, and also due to the advancements in the computational processing power. Though computer-assisted diagnosis (CAD) algorithms in radiology have already complemented the diagnosis by the radiologist, CAD-based algorithms have just started developing and are often used for disease diagnosis and for the prognosis. Nuclear pleomorphism or nuclear atypia in histopathologic images of the breast cancer is a critical prognostic factor in the grading of breast cancer and has been found to have a considerable amount of observer subjectivity and variability when manual scoring is performed. Though analysis and research on histopathologic image has started a decade long, completely automated grading of breast cancer still persists to be a challenge owing to the complexity and large variability of cancer tissues. Thus, active research work is progressing in the area of histopathological image analysis to discover efficient and robust methods for the computer-aided automated grading of cancer.
Another relevant aspect is the lack of standardized publicly available dataset for breast histopathological images. Until recently, majority of the works on histopathologic image analysis of breast cancer were carried out on different datasets that are often small. This gap has been bridged by the release of the BreaKHis dataset and made freely available to the research community by Spanhol et al. [61]. This real life challenging dataset may accelerate research in the breast cancer histopathological image analysis. In [61], they used state-of-the-art texture classification algorithms and came up with an accuracy rate between 80 and 85%, which indicates that there is still advancements to be made in this field for a clinically accepted range of accuracy.
The recently advent deep learning methods are found to be more promising in breast cancer histopathology image classification than handcrafted features. By exploring deeper architectures of neural networks that can extract the inherent features associated with nuclear pleomorphism, more robust models can be developed. Most of the machine learning and deep networks rely on manually annotated datasets for training. The development of a robust deep learning method for breast cancer grading usually requires large amounts of such annotated data. It is often difficult to procure large amount of manually annotated WSIs of breast tumor as the task is tedious and it requires the time and efforts of medically experienced pathologists. A better solution to this and to improve the performance of automated atypia scoring system can be possible through the development of unsupervised learning methods. Unsupervised learning methods can help to overthrow the data annotation phase, thus exploring the histopathology images automatically and accelerating their use. Future work may explore different deeper network architectures, optimization of hyperparameters and also, better strategies for selecting representative patches so as to improve the classification accuracy.
Recent studies have established that often ensembles of classifiers outperform monolithic classifiers [27]. Ensemble of classifiers is a set of several similar classifiers whose individual decisions are combined or aggregated in order to develop a classifier that outperforms each of the individual classifiers in terms of classification accuracy. One of the most dynamic area of research in the field of breast histopathological image classification has been to study techniques for constructing good ensemble of classifiers.
The histopathological image analysis continues to be a challenging one because of the great size of the image data. Moreover, the processing required for the analysis of these images are extensive and analysis of even a single image may take several hours on a single CPU. Because of this, the computational analysis of histopathological images often requires computationally efficient tools and appropriate methods to relieve the problems caused by the size of the images. The processing may be further accelerated through the use of high-performance computing technologies like multi-core processors, graphical processing units (GPUs), or cell processors. GPUs are largely scalable and are becoming more and more powerful and hence histopathological image analysis can be envisaged to be one of the emerging fields to be benefited by the use of GPUs.
Currently, whole-slide digital scanners available on the market are based on the RGB imaging techniques. Spectral imaging or multispectral or hyperspectral imaging can achieve images at different wavelengths instead of ordinary RGB (3-channel) input image [38]. It builds a stack of images with each of the slices representing the images of the same slide acquired by the incident light at varying wavelengths. Spectral imaging is advantageous than RGB systems, because of their capability in analyzing pathological slides colored with different antibodies, which can potentially provide additional significant information to assist in cancer detection. The major challenge with these multichannel-based imaging methods is that these images captured in different wavelengths need to be registered, as there can be misalignments during sequential imaging of the specimen.
H&E stained whole-slide digital imaging system is the traditional imaging applied on histopathology imaging. Many other modalities have also emerged for the imaging of the histological tissues which include point spectroscopy, spectral imaging, and spectroscopic imaging. Registration of these images from various distinct modalities and the multimodal fusion of the data and information encapsulated in them can be a powerful source of information for the diagnosis and the prognosis of cancers. With the availability of highly dense data, a lot of challenges arise in the diagnosis. These may include the extra cost associated with imaging, storage and transmission, processing time, registration, computation, and analysis of this huge amounts of images captured by these multimodal spectral imaging methods. The situation is aggravated with the spectral imaging of whole slides. With the availability of powerful high-performance computing resources, increased storage capacity and transmission bandwidth at surprisingly low costs and recent progresses in the image analysis techniques may help in developing more sophisticated methods for the analysis of huge number of spectral images. However, due to the wide range of imaging modalities and disease complexities, significant results in this area are still unlocked and distinctive challenges and hurdles need to be explored depending on specific applications.
Most of the researchers in the histopathological image analysis field are computing science researchers, and there is a large shortage of pathologists who are experts in computer vision. This can have an adverse effect in the histopathological image based advancements as the field may have to address unique challenges to develop a system with a performance satisfying clinical standards. Hence, it is quite relevant to maintain an active and constant association between the computer vision researchers and the clinical pathologists. It is the pathologists who can better interpret the results of digital analysis with respect to the biological actions and give best suggestions and feedback on the performance of the system. Future studies and research focusing on having some feedback from the pathologies is expected to explore new course of research and help to identify the unexplored areas in the field of histopathological cancer grading.
The adoption of digital histopathological image analysis and diagnosis techniques, for cancer prognosis, can help the pathologists predict the outcome of the disease and the survival chances of a patient. As cancer grading has high correspondence with the prognosis factors, advances in the digital cancer grading can have a significant implication in the prognostic analysis. The increasing availability of the slide scanning equipments in pathological labs along with powerful image analysis algorithms helps in integrating these kinds of quantitative approaches for cancer grading and prognosis in the routine workflow of pathological practices, which can improve the destiny of digital pathology. A complete automated breast cancer diagnosis and grading system for clinical purposes will be a mile stone in the field of medicine, ensuring consistency and repeatability, and eliminating the tediousness of the pathologists.
Conclusion
Over the last few decades, the use of digital analysis techniques for histopathology images has been increasing rapidly, with the introduction of whole-slide imaging and digital slides in pathology labs. The use of computer-aided quantitative analysis of cancer tissues can overcome the inherent problems of lack of repeatability and inter- and intra-observer variability, that can influence the diagnosis of the disease and treatment planning. The breast histopathological image analysis forms a challenging task because of the various variabilities and artifacts occurred by imprecise slide preparation and also due to the complex architecture of the underlying cellular pattern. Hence, the histopathological image analysis techniques developed will have to be robust enough to such variations. This article discussed the techniques and algorithms used for nuclear atypia scoring of breast cancer, challenges in the automated grading, and the future possibilities of computer-aided diagnosis (CAD) systems. This study is a venture to abstract out the recent developments in breast cancer grading and to give an overview on the accuracy and efficiency of different techniques. The study reveals that further improvements are needed in the histopathological image analysis techniques to develop algorithms with performance levels which are acceptable for clinical applications.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Aksac A, Demetrick DJ, Ozyer T, Alhajj R (2019) BrecaHAD: A dataset for breast cancer histopathological annotation and diagnosis. BMC Research Notes 12(1). 10.1186/s13104-019-4121-7 [DOI] [PMC free article] [PubMed]
- 2.Alom MZ, Yakopcic C, Nasrin MS, Taha TM, Asari VK (2019) Breast Cancer Classification from Histopathological Images with Inception Recurrent Residual Convolutional Neural Network. Journal of Digital Imaging. 10.1007/s10278-019-00182-7 [DOI] [PMC free article] [PubMed]
- 3.Araújo T, Aresta G, Castro E, Rouco J, Aguiar P, Eloy C, Polónia A, Campilho A Classification of breast cancer histology images using convolutional neural networks. PloS one 12(6), 2017 [DOI] [PMC free article] [PubMed]
- 4.Bardou D, Zhang K, Ahmad SM. Classification of breast cancer based on histology images using convolutional neural networks. IEEE Access. 2018;6:24,680–24,693. [Google Scholar]
- 5.Basavanhally A, Ganesan S, Feldman M, Shih N, Mies C, Tomaszewski J, Madabhushi A. Multi-field-of-view framework for distinguishing tumor grade in ER+ breast cancer from entire histopathology slides. IEEE Transactions on Biomedical Engineering. 2013;60(8):2089–2099. doi: 10.1109/TBME.2013.2245129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bayramoglu N, Kannala J, Heikkila J (2017) Deep learning for magnification independent breast cancer histopathology image classification. In: Proceedings International Conference on Pattern Recognition, pp 2440–2445. 10.1109/ICPR.2016.7900002
- 7.Beevi KS, Nair MS, Bindu GR. Automatic segmentation of cell nuclei using Krill Herd optimization based multi-thresholding and Localized Active Contour Model. Biocybernetics and Biomedical Engineering. 2016;36(4):584–596. [Google Scholar]
- 8.Bejnordi BE, Zuidhof G, Balkenhol M, Hermsen M, Bult P, van Ginneken B, Karssemeijer N, Litjens G, van der Laak J. Context-aware stacked convolutional neural networks for classification of breast carcinomas in whole-slide histopathology images. Journal of Medical Imaging. 2017;4(4):044,504. doi: 10.1117/1.JMI.4.4.044504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Contesso G, Mouriesse H, Friedman S, Genin J, Sarrazin D, Rouesse J. The importance of histologic grade in long-term prognosis of breast cancer: a study of 1,010 patients, uniformly treated at the Institut Gustave-Roussy. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 1987;5(9):1378–1386. doi: 10.1200/JCO.1987.5.9.1378. [DOI] [PubMed] [Google Scholar]
- 10.Cosatto E, Miller M, Graf HP, Meyer JS (2008) Grading nuclear pleomorphism on histological micrographs. 2008 ICPR 2008 19th International Conference on (August 2016) Pattern Recognition, pp 1–4. 10.1109/ICPR.2008.4761112
- 11.Dalle JR, Leow WK, Racoceanu D, Tutac AE, Putti TC (2008) Automatic breast cancer grading of histopathological images. In: 2008 30th Annual International Conference of the IEEE Engineering in Medicine and Biology Society, pp 3052–3055. 10.1109/IEMBS.2008.4649847 [DOI] [PubMed]
- 12.Dalle Jr, Racoceanu D, Putti TC Nuclear pleomorphism scoring by selective cell nuclei detection. IEEE Workshop on Applications of Computer Vision: 7–8, 2009
- 13.Das A, Nair MS, Peter SD. Sparse representation over learned dictionaries on the riemannian manifold for automated grading of nuclear pleomorphism in breast cancer. IEEE Transactions on Image Processing. 2019;28(3):1248–1260. doi: 10.1109/TIP.2018.2877337. [DOI] [PubMed] [Google Scholar]
- 14.Doyle S, Agner S, Madabhushi A, Feldman M, Tomaszewski J Automated grading of breast cancer histopathology using spectral clustering with textural and architectural image features. In: 2008 5Th IEEE international symposium on biomedical imaging: From nano to macro, Proceedings, ISBI, 2008, pp 496–499. 10.1109/ISBI.2008.4541041
- 15.Easley G, Labate D, Lim WQ. Sparse directional image representations using the discrete shearlet transform. Applied and Computational Harmonic Analysis. 2008;25(1):25–46. [Google Scholar]
- 16.Elmore JG, Longton GM, Carney PA, Geller BM, Onega T, Tosteson ANA, Nelson HD, Pepe MS, Allison KH, Schnitt SJ, O’Malley FP, Weaver DL. Diagnostic Concordance Among Pathologists Interpreting Breast Biopsy Specimens. JAMA. 2015;313(11):1122. doi: 10.1001/jama.2015.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elston CW, Ellis IO (1991) Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: experience from a large study with long term followup, vol 19. 10.1111/j.1365-2559.1991.tb00229.x, arXiv:1011.1669v3 [DOI] [PubMed]
- 18.Faridi P, Danyali H, Helfroush MS, Jahromi MA (2016) Cancerous nuclei detection and scoring in breast cancer histopathological images. arXiv:161201237
- 19.Fawcett T. An introduction to ROC analysis. Pattern Recognition Letters. 2006;27(8):861–874. [Google Scholar]
- 20.Forouzanfar MH, Alexander L, Anderson HR, Bachman VF, Biryukov S, Brauer M, Burnett R, Casey D, Coates MM, Cohen A, et al. Global, regional, and national comparative risk assessment of 79 behavioural, environmental and occupational, and metabolic risks or clusters of risks in 188 countries, 1990–2013: a systematic analysis for the global burden of disease study 2013. The Lancet. 2015;386(10010):2287–2323. doi: 10.1016/S0140-6736(15)00128-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gandomkar Z, Brennan PC, Mello-Thoms C. MudeRN: Multi-category classification of breast histopathological image using deep residual networks. Artificial Intelligence in Medicine. 2018;88:14–24. doi: 10.1016/j.artmed.2018.04.005. [DOI] [PubMed] [Google Scholar]
- 22.Gandomkar Z, Brennan PC, Mello-Thoms C (2019) Computer-Assisted Nuclear Atypia Scoring of Breast Cancer: a Preliminary Study. Journal of Digital Imaging. 10.1007/s10278-019-00181-8 [DOI] [PMC free article] [PubMed]
- 23.Genestie C, Zafrani B, Asselain B, Fourquet A, Rozan S, Validire P, Vincent-Salomon A, Sastre-Garau X. Comparison of the prognostic value of Scarff-Bloom-Richardson and Nottingham histological grades in a series of 825 cases of breast cancer: major importance of the mitotic count as a component of both grading systems. Anticancer Research. 1998;18(1B):571–576. [PubMed] [Google Scholar]
- 24.Ghaznavi F, Evans A, Madabhushi A, Feldman M. Digital imaging in pathology: Whole-slide imaging and beyond. Annual Review of Pathology: Mechanisms of Disease. 2013;8(1):331–359. doi: 10.1146/annurev-pathol-011811-120902. [DOI] [PubMed] [Google Scholar]
- 25.Golatkar A, Anand D, Sethi A (2018) Classification of Breast Cancer Histology Using Deep Learning. In: Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics), vol 10882 LNCS, pp 837–844. 10.1007/978-3-319-93000-8_95
- 26.Guo Y, Dong H, Song F, Zhu C, Liu J (2018) Breast Cancer Histology Image Classification Based on Deep Neural Networks. In: Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics), vol 10882 LNCS, pp 827–836. 10.1007/978-3-319-93000-8-94, 1803.04054
- 27.Gurcan MN, Boucheron LE, Can A, Madabhushi A, Rajpoot NM, Yener B. Histopathological image analysis: a review. IEEE Reviews in Biomedical Engineering. 2009;2:147–171. doi: 10.1109/RBME.2009.2034865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hammond MEH, Hayes DF, Dowsett M, Allred DC, Hagerty KL, Badve S, Fitzgibbons PL, Francis G, Goldstein NS, Hayes M, et al. American society of clinical oncology/college of american pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer (unabridged version) Archives of pathology & laboratory medicine. 2010;134(7):e48–e72. doi: 10.5858/134.7.e48. [DOI] [PubMed] [Google Scholar]
- 29.Han Z, Wei B, Zheng Y, Yin Y, Li K, Li S (2017) Breast Cancer Multi-classification from Histopathological Images with Structured Deep Learning Model. Scientific Reports 7(1). 10.1038/s41598-017-04075-z [DOI] [PMC free article] [PubMed]
- 30.Harris J, Lippman M, Morrow M, Kent osborne C (2014) Diseases of the breast, 5th edition
- 31.Hossin M, Sulaiman M. A review on evaluation metrics for data classification evaluations. International Journal of Data Mining &, Knowledge Management Process. 2015;5(2):01–11. [Google Scholar]
- 32.Huang CH, Veillard A, Roux L, Lomėnie N, Racoceanu D. Time-efficient sparse analysis of histopathological whole slide images. Computerized Medical Imaging and Graphics. 2011;35(7-8):579–591. doi: 10.1016/j.compmedimag.2010.11.009. [DOI] [PubMed] [Google Scholar]
- 33.Irshad H, Veillard A, Roux L, Racoceanu D. Methods for nuclei detection, segmentation, and classification in digital histopathology: a review-current status and future potential. IEEE Reviews in Biomedical Engineering. 2014;7:97–114. doi: 10.1109/RBME.2013.2295804. [DOI] [PubMed] [Google Scholar]
- 34.Jannesari M, Habibzadeh M, Aboulkheyr H, Khosravi P, Elemento O, Totonchi M, Hajirasouliha I (2019) Breast cancer histopathological image classification: a deep learning approach. In: Proceedings - 2018 IEEE International Conference on Bioinformatics and Biomedicine, BIBM 2018, pp 2405–2412. 10.1109/BIBM.2018.8621307
- 35.Jiang Y, Chen L, Zhang H, Xiao X (2019) Breast cancer histopathological image classification using convolutional neural networks with small SE-resnet module. PLoS ONE 14(3). 10.1371/journal.pone.0214587 [DOI] [PMC free article] [PubMed]
- 36.Jimenez-deltaro O, Otlora S, Andersson M, Eurén K, Hedlund M, Rousson M, Müller H, Atzori M (2018) Analysis of histopathology images: From traditional machine learning to deep learning. In: Biomedical Texture Analysis, Elsevier, pp 281–314
- 37.Jovanovic J (2016) Classification. http://ai.fon.bg.ac.rs/wp-content/uploads/2016/10/Classification-basic-concepts.pdf
- 38.Kårsnäs A (2014) Image analysis methods and tools for digital histopathology applications relevant to breast cancer diagnosis. PhD thesis
- 39.Khan AM, Rajpoot N, Treanor D, Magee D. A nonlinear mapping approach to stain normalization in digital histopathology images using image-specific color deconvolution. IEEE Transactions on Biomedical Engineering. 2014;61(6):1729–1738. doi: 10.1109/TBME.2014.2303294. [DOI] [PubMed] [Google Scholar]
- 40.Khan AM, Sirinukunwattana K, Rajpoot N. A global covariance descriptor for nuclear atypia scoring in breast histopathology images. IEEE Journal of Biomedical and Health Informatics. 2015;19(5):1637–1647. doi: 10.1109/JBHI.2015.2447008. [DOI] [PubMed] [Google Scholar]
- 41.Kumar R, Srivastava R, Srivastava S. Detection and classification of cancer from microscopic biopsy images using clinically significant and biologically interpretable features. Journal of Medical Engineering. 2015;2015(2015):1–14. doi: 10.1155/2015/457906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lu C, Ji M, Ma Z, Mandal M. Automated image analysis of nuclear atypia in high-power field histopathological image. Journal of Microscopy. 2015;258(3):233–240. doi: 10.1111/jmi.12237. [DOI] [PubMed] [Google Scholar]
- 43.Lyon HO, De Leenheer AP, Horobin RW, lambert WE, schulte EK, Van Liedekerke B, Wittekind DH (1994) Standardization of reagents and methods used in cytological and histological practice with emphasis on dyes, stains and chromogenic reagents. 10.1007/BF00158587 [DOI] [PubMed]
- 44.Macenko M, Niethammer M, Marron JS, Borland D, Woosley JT, Guan X, Schmitt C, Thomas NE (2009) A method for normalizing histology slides for quantitative analysis. In: Proceedings - 2009 IEEE International Symposium on Biomedical Imaging: From Nano to Macro, ISBI 2009, pp 1107–1110. 10.1109/ISBI.2009.5193250
- 45.Malvia S, Bagadi SA, Dubey US, Saxena S Epidemiology of breast cancer in indian women 13(4), 289–295, 2017. 10.1111/ajco.12661 [DOI] [PubMed]
- 46.Maqlin P, Thamburaj R, Mammen JJ, Manipadam MT (2015) Automated nuclear pleomorphism scoring in breast cancer histopathology images using deep neural networks. In: Lecture notes in computer science (including subseries lecture notes in artificial intelligence and lecture notes in bioinformatics), vol 9468, pp 269–276. 10.1007/978.3.319.26832.3.26
- 47.Moncayo R, Romo-Bucheli D, Romero E (2015) A grading strategy for nuclear pleomorphism in histopathological breast cancer images using a bag of features (bof). In: Lecture notes in computer science (including subseries lecture notes in artificial intelligence and lecture notes in bioinformatics), vol 9423, pp 75–82. 10.1007/978.3.319.25751.8.10
- 48.Nahid AA, Mehrabi MA, Kong Y (2018) Histopathological breast cancer image classification by deep neural network techniques guided by local clustering. BioMed research international 2018 [DOI] [PMC free article] [PubMed]
- 49.Naik S, Doyle S, Agner S, Madabhushi A, Feldman M, Tomaszewski J (2008) Automated gland and nuclei segmentation for grading of prostate and breast cancer histopathology. In: 2008 5Th IEEE international symposium on biomedical imaging: From nano to macro, Proceedings, ISBI, pp 284–287. 10.1109/ISBI.2008.4540988
- 50.Nejad EM, Affendey LS, Latip RB, Bin Ishak I (2017) Classification of Histopathology Images of Breast into Benign and Malignant using a Single-layer Convolutional Neural Network. In: Proceedings of the International Conference on Imaging, Signal Processing and Communication - ICISPC, vol 2017, pp 50–53. 10.1145/3132300.3132331
- 51.Niethammer M, Borland D, Marron J, Woosley JT, Thomas NE (2010) Appearance normalization of histology slides. In: MLMI, Springer, pp 58–66 [DOI] [PMC free article] [PubMed]
- 52.Ojansivu V, Linder N, Rahtu E, Pietikäinen M, Lundin M, Joensuu H, Lundin J. Automated classification of breast cancer morphology in histopathological images. Diagnostic Pathology. 2013;8(1):S29. [Google Scholar]
- 53.Petushi S, Katsinis C, Coward C, Garcia F, Tozeren A (2004) Automated identification of microstructures on histology slides. In: 2004 IEEE International Symposium on Biomedical imaging: Nano to macro, IEEE, pp 424–427
- 54.Petushi S, Garcia FU, Haber MM, Katsinis C, Tozeren A. Large-scale computations on histology images reveal grade-differentiating parameters for breast cancer. BMC Medical Imaging. 2006;6(1):14. doi: 10.1186/1471-2342-6-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rakhlin A, Shvets A, Iglovikov V, Kalinin AA (2018) Deep Convolutional Neural Networks for Breast Cancer Histology Image Analysis. In: Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics), vol 10882 LNCS, pp 737–744. 10.1007/978-3-319-93000-8-83, 1802.00752
- 56.Rezaeilouyeh H, Mollahosseini A, Mohammad MH. Microscopic medical image classification framework via deep learning and shearlet transform. Journal of Medical Imaging. 2016;3(4):044,501. doi: 10.1117/1.JMI.3.4.044501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rolls G (2010) Microtomy and Paraffin Section Preparation. Scientia Leica Microsystems’ Education Series, pp 32, https://www.leica-microsystems.com
- 58.Roux L, Racoceanu D, Lomėnie N, Kulikova M, Irshad H, Klossa J, Capron F, Genestie C, Naour G, Gurcan M. Mitosis detection in breast cancer histological images an ICPR 2012 contest. Journal of Pathology Informatics. 2013;4(1):8. doi: 10.4103/2153-3539.112693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ruifrok AC, Johnston DA. Quantification of histochemical staining by color deconvolution. Analytical and Quantitative Cytology and Histology. 2001;23(4):291–299. [PubMed] [Google Scholar]
- 60.Salahuddin T, Haouari F, Islam F, Ali R, Al-Rasbi S, Aboueata N, Rezk E, Jaoua A Breast cancer image classification using pattern-based Hyper Conceptual Sampling method. Informatics in Medicine Unlocked, pp 1–10, 2018. 10.1016/j.imu.2018.07.002
- 61.Spanhol FA, Oliveira LS, Petitjean C, Heutte L. A dataset for breast cancer histopathological image classification. IEEE Transactions on Biomedical Engineering. 2016;63(7):1455–1462. doi: 10.1109/TBME.2015.2496264. [DOI] [PubMed] [Google Scholar]
- 62.Spanhol FA, Oliveira LS, Cavalin PR, Petitjean C, Heutte L (2017) Deep features for breast cancer histopathological image classification. In: 2017 IEEE International Conference on Systems, Man, and Cybernetics (SMC), IEEE, pp 1868– 1873
- 63.Stierer M, Rosen H, Weber R. Nuclear pleomorphism, a strong prognostic factor in axillary node-negative small invasive breast cancer. Breast Cancer Research and Treatment. 1991;20(2):109–116. doi: 10.1007/BF01834640. [DOI] [PubMed] [Google Scholar]
- 64.Teot LA, Sposto R, Khayat A, Qualman S, Reaman G, Parham D. The problems and promise of central pathology review: development of a standardized procedure for the Children’s Oncology Group. Pediatric and developmental pathology : the official journal of the Society for Pediatric Pathology and the Paediatric Pathology Society. 2007;10(3):199–207. doi: 10.2350/06-06-0121.1. [DOI] [PubMed] [Google Scholar]
- 65.Vang YS, Chen Z, Xie X (2018) Deep learning framework for multi-class breast cancer histology image classification. In: International Conference Image Analysis and Recognition, Springer, pp 914–922
- 66.Vesal S, Ravikumar N, Davari AA, Ellmann S, Maier A (2018) Classification of Breast Cancer Histology Images Using Transfer Learning. In: Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics), vol 10882 LNCS, pp 812–819. 10.1007/978-3-319-93000-8-92, 1802.09424
- 67.Veta M, Kornegoor R, Huisman A, Verschuur-Maes AHJ, Viergever MA, Pluim JPW, van Diest PJ. Prognostic value of automatically extracted nuclear morphometric features in whole slide images of male breast cancer. Modern Pathology. 2012;25(12):1559–1565. doi: 10.1038/modpathol.2012.126. [DOI] [PubMed] [Google Scholar]
- 68.Veta M, van Diest PJ, Willems SM, Wang H, Madabhushi A, Cruz-Roa A, Gonzalez F, Larsen AB, Vestergaard JS, Dahl AB, Cirean DC, Schmidhuber J, Giusti A, Gambardella LM, Tek FB, Walter T, Wang CW, Kondo S, Matuszewski BJ, Precioso F, Snell V, Kittler J, de Campos TE, Khan AM, Rajpoot NM, Arkoumani E, Lacle MM, Viergever MA, Pluim JP. Assessment of algorithms for mitosis detection in breast cancer histopathology images. Medical Image Analysis. 2015;20(1):237–248. doi: 10.1016/j.media.2014.11.010. [DOI] [PubMed] [Google Scholar]
- 69.Wan T, Cao J, Chen J, Qin Z. Automated grading of breast cancer histopathology using cascaded ensemble with combination of multi-level image features. Neurocomputing. 2017;229:34–44. [Google Scholar]
- 70.Wei B, Han Z, He X, Yin Y (2017) Deep learning model based breast cancer histopathological image classification. In: 2017 IEEE 2nd International Conference on Cloud Computing and Big Data Analysis (ICCCBDA), pp 348–353. 10.1109/ICCCBDA.2017.7951937
- 71.Wetzel AW, John RGI, Beckstead JA, Feineigle PA, Hauser CR, Palmieri FA Jr (2006) System for creating microscopic digital montage images. US Patent 7,155,049
- 72.Weyn B, Van De Wouwer G, Van Daele A, Scheunders P, Van Dyck D, Van Marck E, Jacob W: Automated breast tumor diagnosis and grading based on wavelet chromatin texture description. Cytometry 33 (1): 32–40, 1998. 10.1002/(SICI)1097-0320(19980901)33:1.32::AID-CYTO4.3.0.CO;2-D [DOI] [PubMed]
- 73.Wolberg WH, Street WN, Heisey DM, Mangasarian OL. Computer-derived nuclear ”grade” and breast cancer prognosis. Analytical and Quantitative Cytology and Histology. 1995;17(4):257–64. [PubMed] [Google Scholar]
- 74.Wolff AC, Hammond MEH, Schwartz JN, Hagerty KL, Allred DC, Cote RJ, Dowsett M, Fitzgibbons PL, Hanna WM, Langer A, et al. American society of clinical oncology/college of american pathologists guideline recommendations for human epidermal growth factor receptor 2 testing in breast cancer. Journal of Clinical Oncology. 2006;25(1):118–145. doi: 10.1200/JCO.2006.09.2775. [DOI] [PubMed] [Google Scholar]
- 75.Wollmann T, Rohr K (2017) Automatic breast cancer grading in lymph nodes using a deep neural network. arXiv:170707565
- 76.Xu J, Zhou C, Lang B, Liu Q (2017) Deep learning for histopathological image analysis: Towards computerized diagnosis on cancers. In: Advances in Computer Vision and Pattern Recognition, pp 73–95, 10.1007/978-3-319-42999-1-6