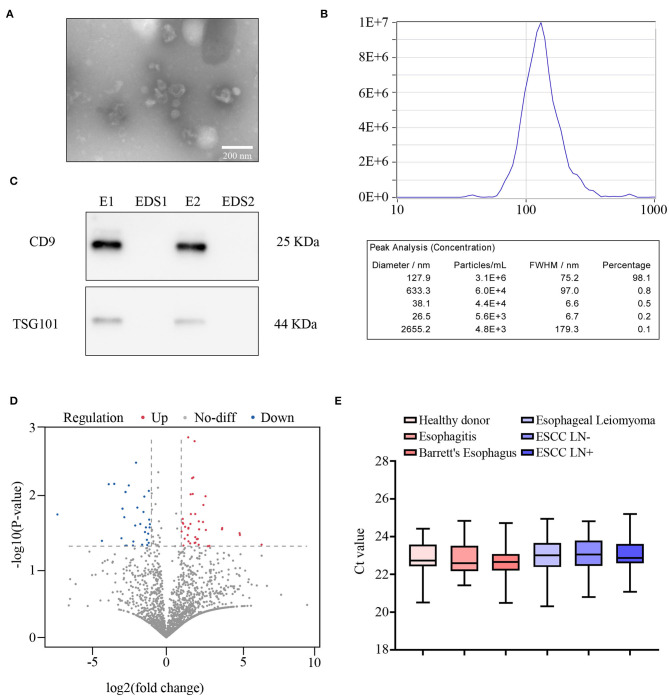
Figure 2.
Primary data collection and global screening of microRNA (miRNA) expression to predict lymph node (LN) metastasis. (A) The morphology of exosomes was identified using transmission electron microscopy (TEM). Scale bar, 100 nm. (B) Size distribution and concentration of exosomes. (C) Western blot assay of CD9 and TSG101 in exosome samples (E) and exosome-depleted supernatants (EDSs). (D) Volcano plot showing the differentially expressed miRNAs from next-generation sequencing (NGS) analysis. (E) Comparison of internal control exosomal miR-16 expression among HD (n = 38), reflux esophagitis (n = 30), Barrett's esophagus (n = 18), esophageal leiomyoma (n = 15), LN– (n = 30), and LN+ (n = 30) detected by qRT-PCR.