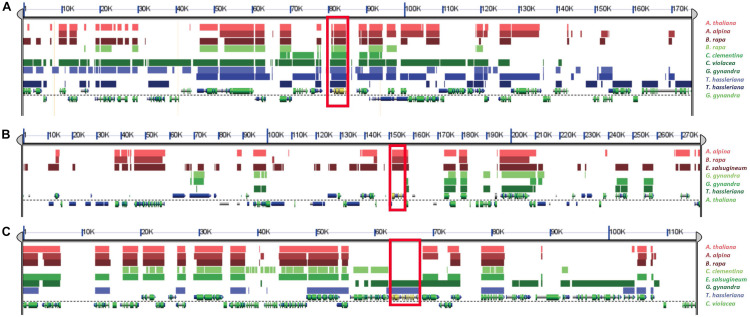
FIGURE 5.
Syntenic relationships of β-NAD-ME and α-NAD-ME genes across Cleomaceae, Brassicaceae and outgroup genomes using SynFind and GEvo analysis. (A) A search for syntenic regions based on the β-NAD-ME1 gene in G. gynandra (as reference region) identified syntenic homologs in all species (shown in red box), including the outgroup species C. clementina. Also, all duplicated copies found by phylogenetic analysis of β-NAD-ME, for example in B. rapa and T. hassleriana, were also syntenic which supports that they are derived from ancient polyploidy events. (B) A search for syntenic regions based on the A. thaliana (Brassicaceae) copy of α-NAD-ME identified only single copy syntenic homologs (shown within red box) in other Brassicaceae species (A. alpina, E. salsugineum, and B. rapa) but not with Cleomaceae species (C. violacea, G. gynandra, and T. hassleriana) nor the outgroup species C. clementenia. Similarly, (C) A search for syntenic homologs of the C. violacea α-NAD-ME gene (shown within red box) found them only in the other Cleomaceae species (G. gynandra and T. hassleriana) but not with Brassicaceae or the outgroup species. These combined results (B, C) suggest independent translocations of α-NAD-ME genes in the Brassicaceae and Cleomaceae to new genomic contexts.