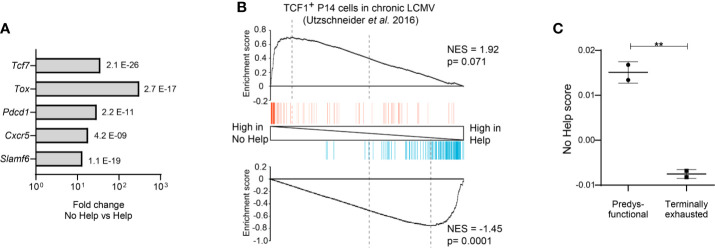
Figure 2.
Predysfunctional TCF-1+ CD8+ T cells in a chronic LCMV infection model display a gene expression signature characteristic of helpless antigen-specific CD8+ T cells in a vaccination model. The transcriptional “No Help” signature was determined by differential gene expression (False discovery rate (FDR) < 0.01) of antigen-specific CTLs raised in No Help versus Help vaccination settings (GEO database GSE89665) (3). (A) Differential expression of selected genes characteristic of predysfunctional TCF-1+CD8+ T cells (Table 1) in No Help versus Help settings. FDR is depicted per gene. (B) GSEA of the top 200 upregulated (red)- or downregulated (blue) genes from TCF1+ versus TCF1− virus-specific CD8+ T cells in chronic LCMV infection (9) within the gene expression profiles of CD8+ T cells from the No Help versus Help vaccination settings. NES, normalized enrichment score. (C) No Help score in predysfunctional TCF-1+TIM3− and terminally exhausted TCF-1−TIM3+ CD8+ T cells from a setting of chronic LCMV infection (GEO database GSE122713) (7). The No Help score was calculated as the difference of correlations in gene expression between a given population and No Help or Help vaccination settings. A higher No-Help score indicates greater transcriptional similarity to helpless CD8+ T cells. **P < 0.01 by Student’s t-test.