Figure 2.
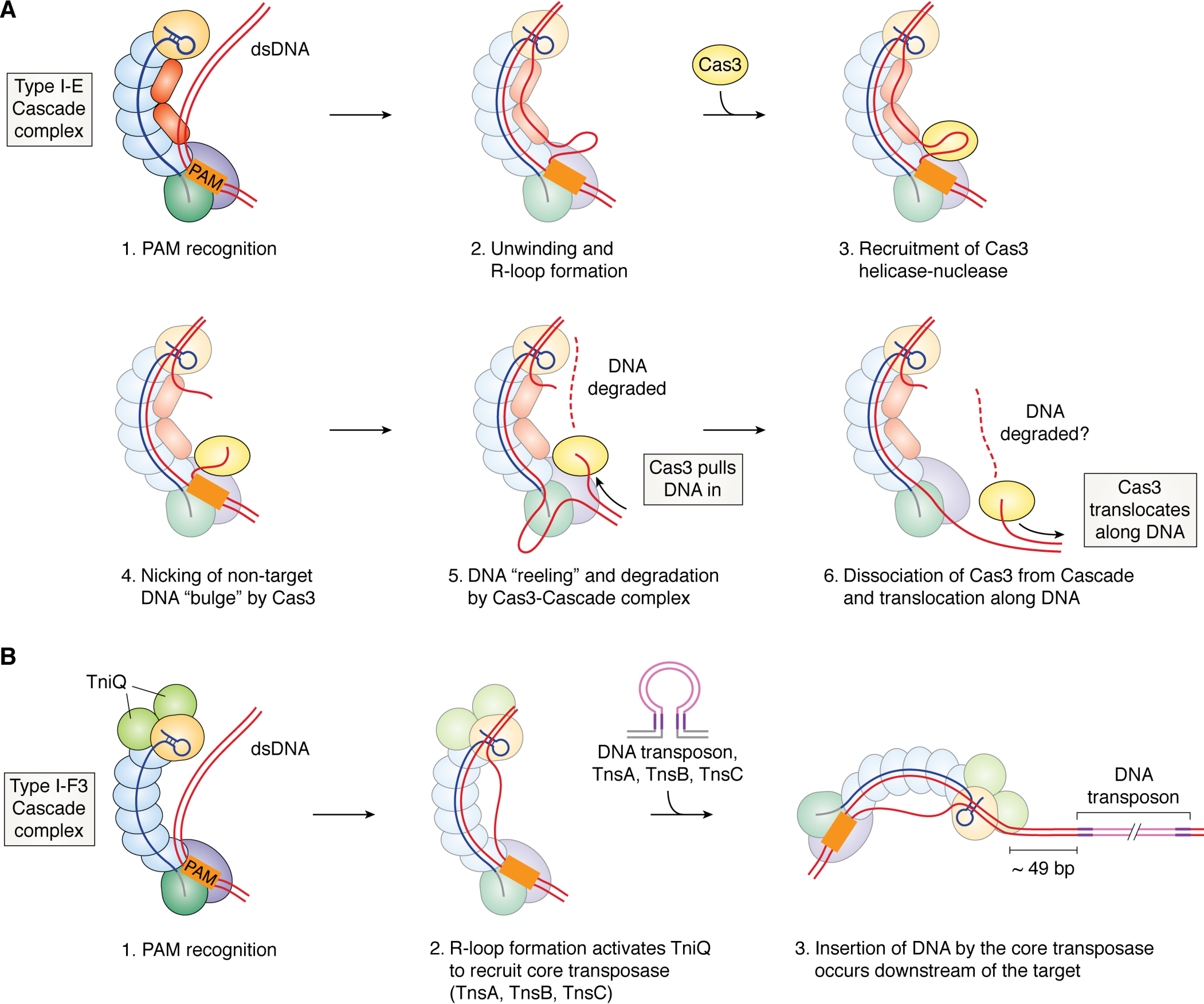
Type I CRISPR-Cas interference mechanism. A, target binding and degradation by the Type I-E Cascade complex. 1, recognition of the PAM (shown as an orange rectangle) by Cas8 leads to DNA bending and initiation of unwinding; 2, hybridization of the crRNA with the target strand of DNA leads to displacement of the nontarget DNA strand and formation of an R-loop; 3, a conformational change in Cascade that accompanies R-loop formation leads to recruitment of Cas3, a helicase-nuclease protein, to a small bulge in the nontarget strand; 4, the nontarget strand bulge is cleaved by Cas3, and the ssDNA is loaded into the helicase; 5, Cas3's helicase domain unwinds DNA upstream of the PAM, and “reels in” ssDNA toward its nuclease active site; 6, Cas3 dissociates from Cascade and continues to translocate; whether or not degradation occurs during translocation is unclear. B, cooperation of Type I-F3 Cascade with a Tn7 transposase to mediate RNA-guided DNA insertion. Recognition of the PAM (1) and base-pairing with a complementary DNA sequence (2) leads to R-loop formation, which activates the TniQ dimer to recruit the core transposase (TnsA, TnsB, TnsC) and transposon DNA with paired ends (violet lines) to the Cascade-bound target site. 3, integration of the transposon's DNA cargo (pink lines) occurs ∼49 bp downstream of the DNA target. In all panels, individual Cas proteins are colored as in Fig. 1, but with colors muted for greater clarity of the nucleic acid strands.
