Fig. 1.
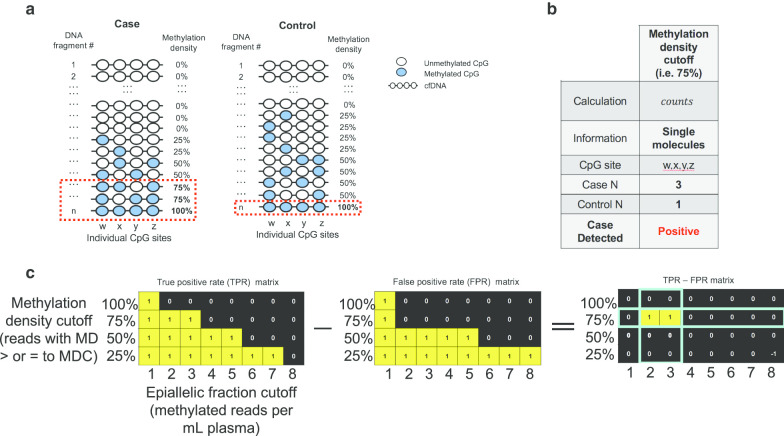
Schematic of the methylation density binary classifier. a Hypothetical dataset of methylation data from a single case and control cfDNA sample containing heterogeneously-methylated DNA b Table characterizing the use of a methylation density classifier cutoff. c Schematic illustrating the EpiClass procedure for determining optimal methylation density and epiallelic fraction cutoffs. The optimal methylation density cutoff is 75%, and the epiallelic fraction cutoff is either 2 or 3. Thus, a sample in this example is called positive if it has 2 or more epialleles that exhibit methylation densities of 75% or higher