Figure 2.
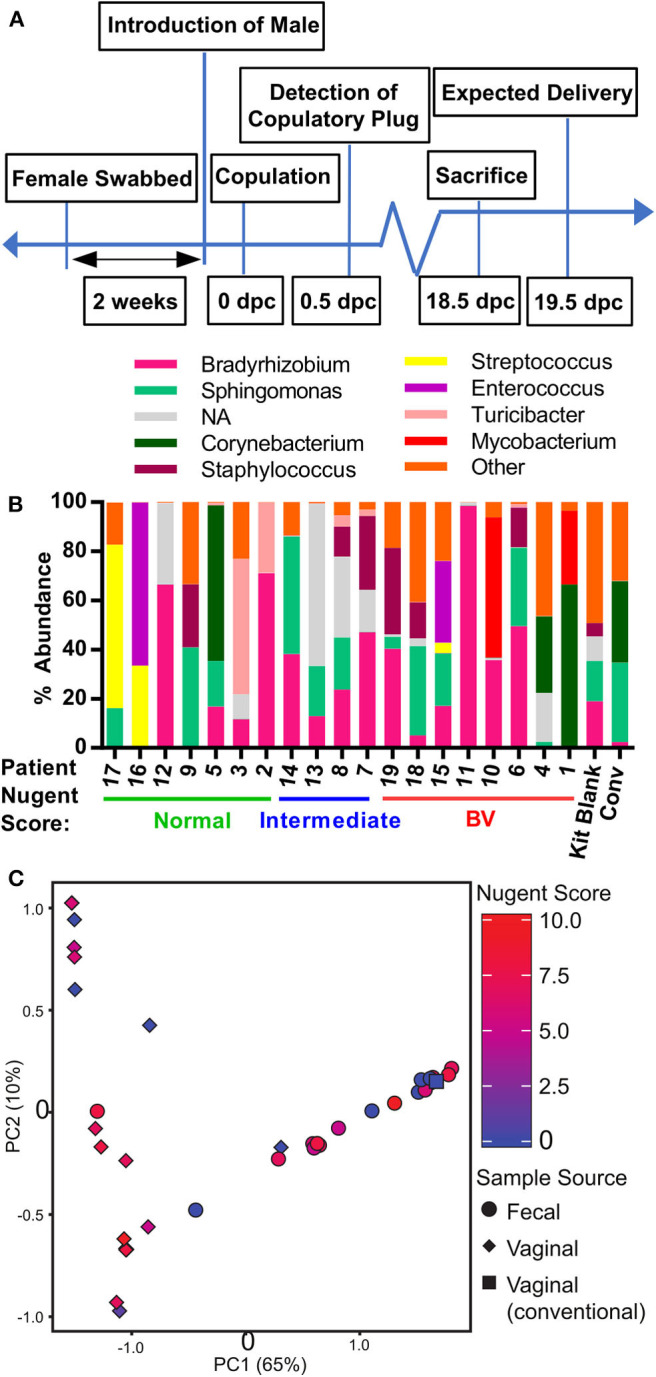
Generation of human microbiota-associated (HMA) mice harboring the microbiota collected from the vaginal tract of pregnant women with bacterial vaginosis. (A) Graphical depiction of experimental approach to generate human microbiota-associated (HMA) mice harboring the microbiota collected from the vaginal tract of pregnant patients described in Table 1. Swabs were collected from the vaginal tract and immediately transported to the Emory Gnotobiotic Animal Core (EGAC). The vaginal tract of female germ-free C57BL/6 were inoculated by physically wiping the swab on the vaginal opening of the mouse. Mice were then housed in Tecniplast ISOcageP Bioexclusion cages for the microbiota to colonize. After 2 weeks, a male germ-free mouse was introduced to the HMA female mouse and conception monitored. On 18.5 dpc (days post-coitum), pregnant female mice were sacrificed under sterile conditions for sample collection and analysis. (B) Relative abundance of the bacterial genera detected via 16S analysis in the vaginal tract of HMA mice on 18.5 dpc. Data represents the top 10 bacterial genera detected and each stacked column represents one mouse. Data is separated by BV status of the corresponding human donor. The total read count for each sample is provided at the top of each bar. NA (not assigned) refers to sequences that were unclassifiable at this taxonomic level. (C) Principal Component Analysis (PCA) plot depicting the beta-diversity of the microbiota community structure within the vaginal tract and gastrointestinal tract of HMA mice on 18.5 dpc. Symbols are colored by the Nugent score of the corresponding human donor.
