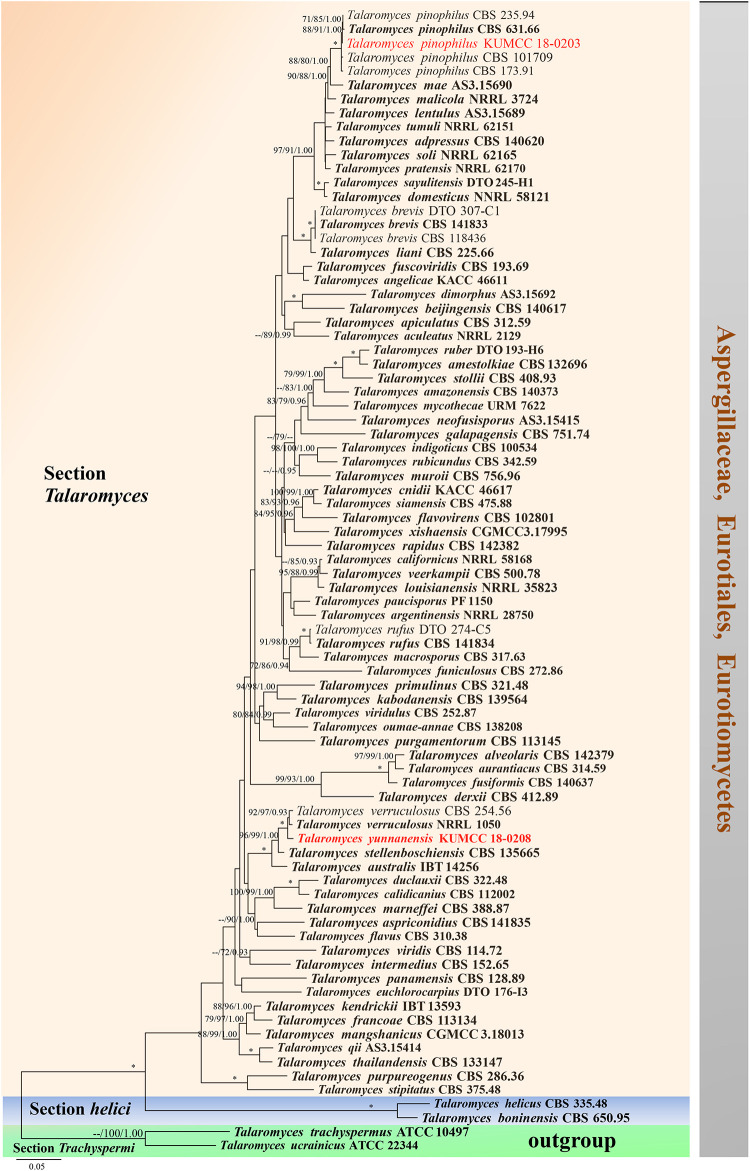
FIGURE 15.
Phylogram generated from maximum likelihood analysis based on combined ITS, BenA, CaM, and RPB2 sequence data. Eighty-two sequences are included in the combined analysis, which consisted of 2858 characters including alignment gaps. The tree is rooted to Talaromyces trachyspermus (ATCC 10497) and T. ucrainicus (ATCC 22344). The MP analysis for the combined dataset had 857 parsimony informative, 1552 constant, 176 parsimony uninformative characters and yielded 46 most parsimonious trees. The best-fit model SYM + I + G was selected for CaM, GTR + I + G for ITS and HKY + I + G for RPB2 and BenA in BI analysis. Maximum parsimony and maximum likelihood bootstrap values ≥ 70% and Bayesian posterior probabilities ≥ 0.90 (MPBS/MLBS/BYPP) are indicated at the nodes. Branches with 100% MPBS, 100% MLBS and 1.00 BYPP values are indicated as an asterisk (∗). The ex-type strains are bolded black, and the new isolates are in red.