Figure 2.
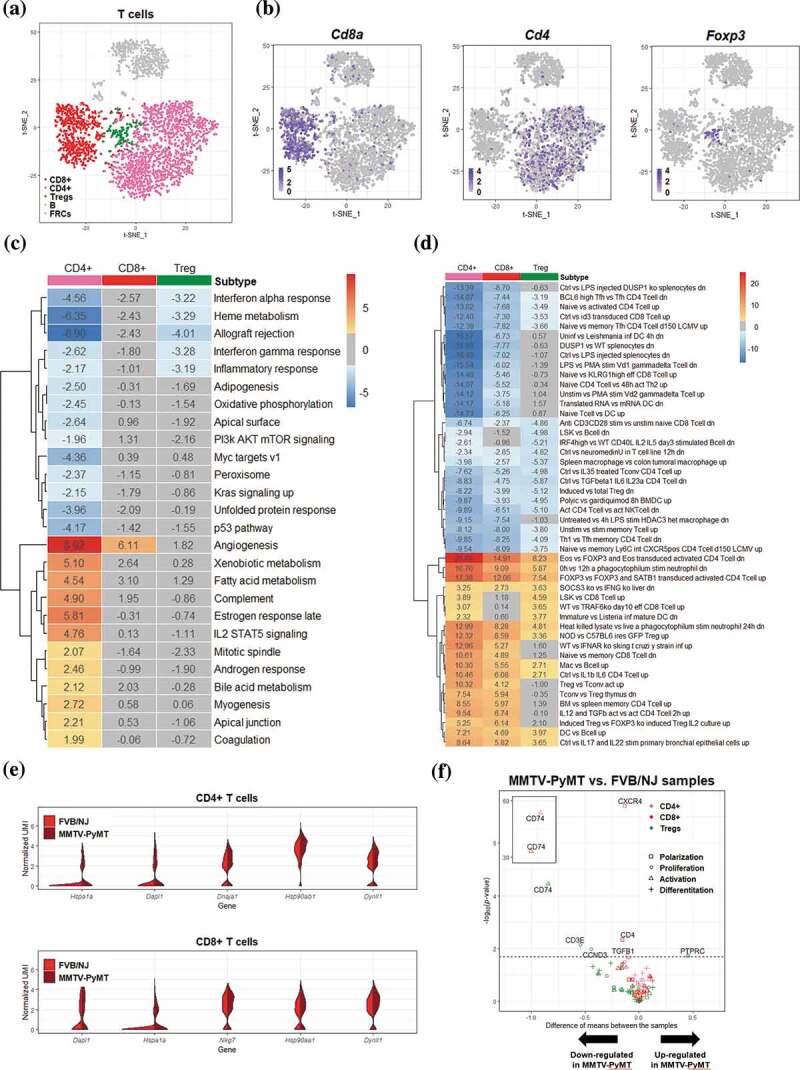
Differential transcriptomic analysis of T cells between the FVB/NJ and MMTV-PyMT samples
(A-B) T cells in tSNE plots, color-coded by the annotated cell types (A) and by various marker genes of T cells (B). (C-D) Heatmap representation of the results of GSVA based on hallmark gene sets (C) and immunologic signatures C7 (D). The value in each box represents the difference between the enrichment scores (ESs) of a gen set in a cell type in the FVB/NJ and in MMTV-PyMT samples, and a positve value represents upregulation of a gene set/pathway in PyMT sample. Boxes colored in gray represents q value > 0.1. Only gene sets with q value< ?? In at least one T cell subtype were presented. (E) Violin plots showing the distribution of gene expression levels of top 5 upregulated genes in CD4+ and CD8 + T cells (FVB/NJ vs MMTV-PyMT). (F) Vocano plots showing differential expression of selected cell status marker gene in CD4+, CD8+ and Treg cell populationss between FVB/NJ and MMTV-PyMT samples.
