Figure 2.
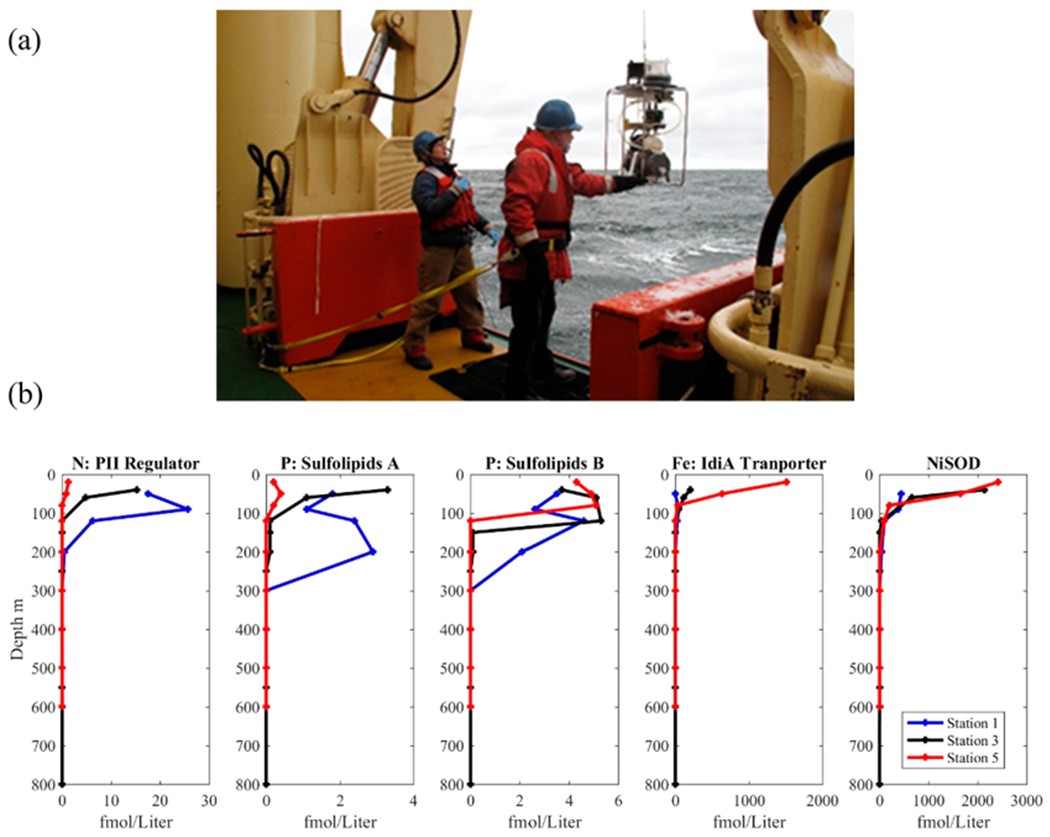
(a) Collection of ocean metaproteomic samples by in situ underwater McLane pump sampler as deployed in Terra Nova Bay of the Ross Sea in Antarctica aboard the icebreaker R/V Palmer to capture the microbial and algal communities as well as larger sinking particles by filtration of several hundreds of liters. (b) Example vertical distributions of three microbial proteins in the Equatorial Pacific Ocean using targeted metaproteomics that are biomarkers of nitrogen (N), phosphorus (P), iron (Fe) nutrient stress, and nickel (Ni) biogeochemical cycling (data from Saito et al., 2014, https://www.bco-dmo.org/dataset/646115). Proteins shown include the nitrogen PII regulator protein from Prochlorococcus (sequence VNSVIDAIAEAAK), the sulfolipid biosynthesis protein from Prochlorococcus (NEAVENDLIVDNK), UDP sulfolipid biosynthesis protein from multiple taxa (FDYDGDYGTVLNR), the IdiA iron transporter from Prochlorococcus (SPYNQSLVANQIVNK), and the nickel superoxide dismutase enzyme from Prochlorococcus and Synechococcus (VAAEAVLSMTK). Taxonomic assignments determined using METATRYP.14
