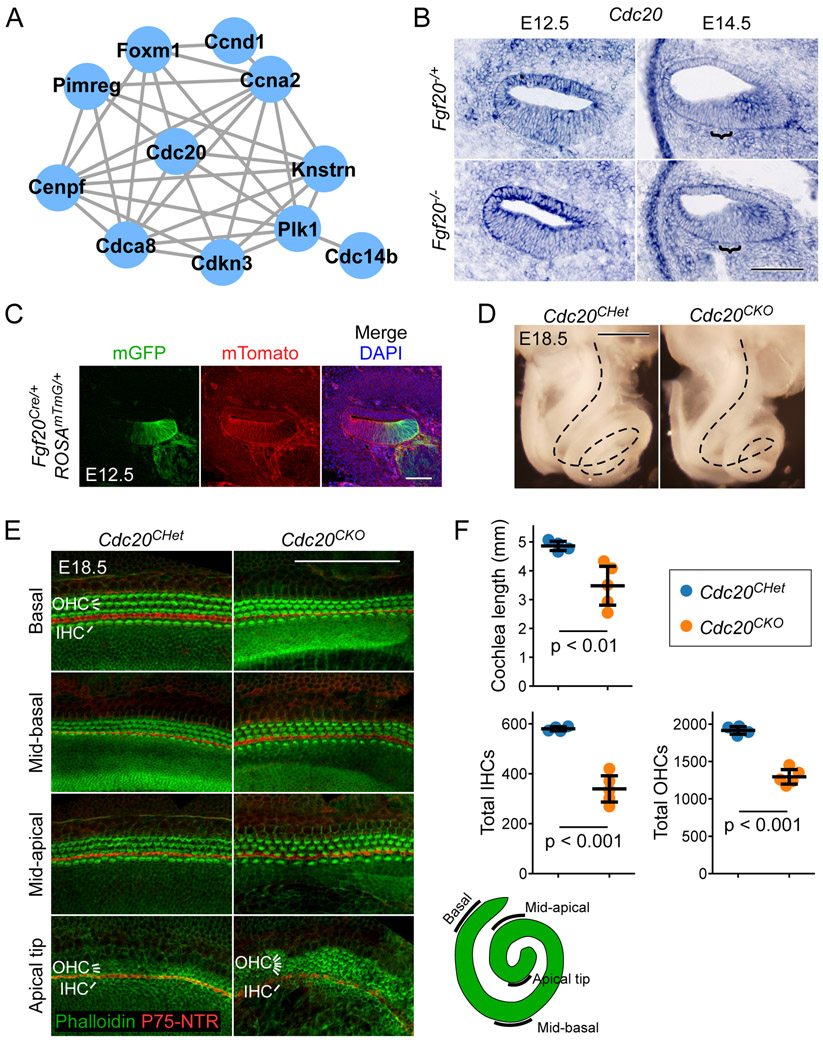
Figure 5. Conditional deletion of Cdc20 with Fgf20Cre resulted in a shorter cochlea.
(A) The largest protein-protein interaction network identified via the STRING database consisted of genes involved in cell cycle regulation. Lines represent known and predicted protein-protein interactions of high or very high confidence (minimum required interaction score = 0.700).
(B) RNA in situ hybridization for Cdc20 on sections through the middle turn of E12.5 and E14.5 Fgf20−/+ (Fgf20Cre/+) and Fgf20−/− (Fgf20Cre/βgal) cochlear ducts. Bracket, prosensory domain. Scale bar, 100 μm.
(C) Section through the middle turn of E12.5 Fgf20Cre/+;ROSAmTmG/+ cochlear duct. Cells of the Fgf20Cre lineage express mGFP (mG, green); non-lineage cells express mTomato (mT, red). DAPI, nuclei (blue); scale bar, 100 μm.
(D) Dissected inner ears from E18.5 Cdc20CHet (Fgf20Cre/+;Cdc20flox/+) and Cdc20CKO (Fgf20Cre/+;Cdc20flox/flox) embryos with the otic capsule removed to reveal the coiled cochlea (dotted lines). Scale bar, 0.5 mm.
(E) Whole mount cochlea from E18.5 Cdc20CHet and Cdc20CKO embryos showing one row of inner hair cells (IHC) and three rows of outer hair cells (OHC) marked by phalloidin (green) and separated by inner pillar cells (p75NTR, red). Representative regions from the basal, mid-basal, and mid-apical turns, and apical tip of the cochlea are shown. See schematic showing locations of the turns of the cochlea. At the apical tip, four or more rows of OHCs were frequently observed in Cdc20CHet cochleae. Scale bar, 100 μm.
(F) Quantification of cochlea length and total number of inner and outer hair cells (IHCs and OHCs) in E18.5 Cdc20CHet (n = 4) and Cdc20CKO (n = 5) cochleae. Error bars represent mean ± std. Results were analyzed by Student’s t-test; p-values are shown.