Figure 4. 5hmC modification of nascent strand causes degradation of stalled replication forks.
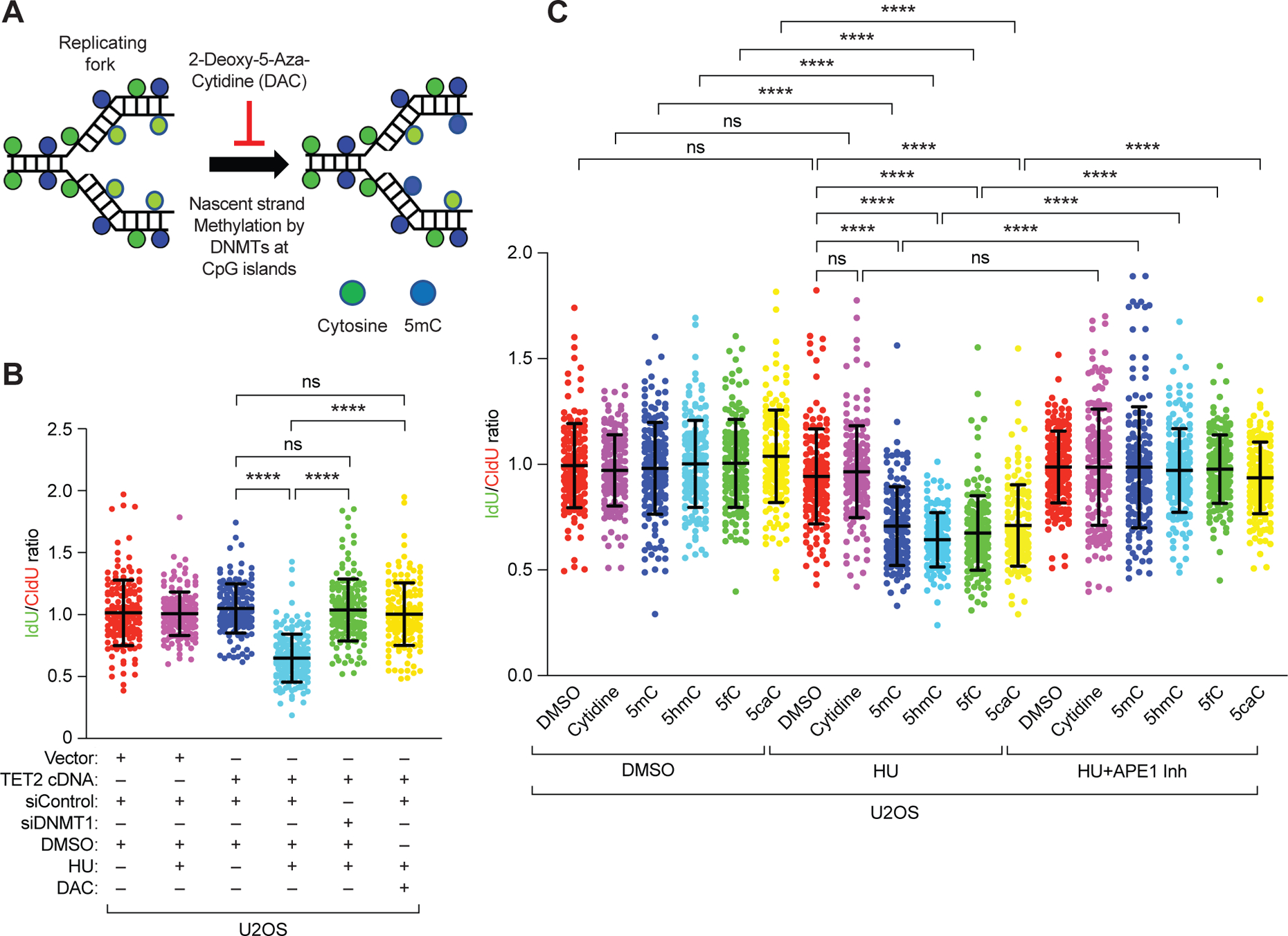
(A) Schematic of the hypothesis that inhibiting DNMT1-mediated methylation of DNA in replication forks with 2-deoxy-5-aza-cytidine (DAC) during DNA fiber analysis will inhibit methylation of nascent DNA strand. (B) Scatter plot of the ratio of IdU:CldU (green:red) in DNA fibers of U2OS cells after TET2 overexpression and DNMT1 knockdown or its inhibition by DAC and in response to either DMSO or HU. (C) Scatter plots showing ratio of IdU:CldU DNA fibers in U2OS cells after treatment with cytidine or modified cytidine residues (1 μM), in the presence of either DMSO, HU, or HU and APE1 inhibitor (600 nM). Data are mean ±SD from n = 2, N = ~150 DNA fibers. ns P≥0.05, and **** P≤0.0001 by Mann-Whitney test.
