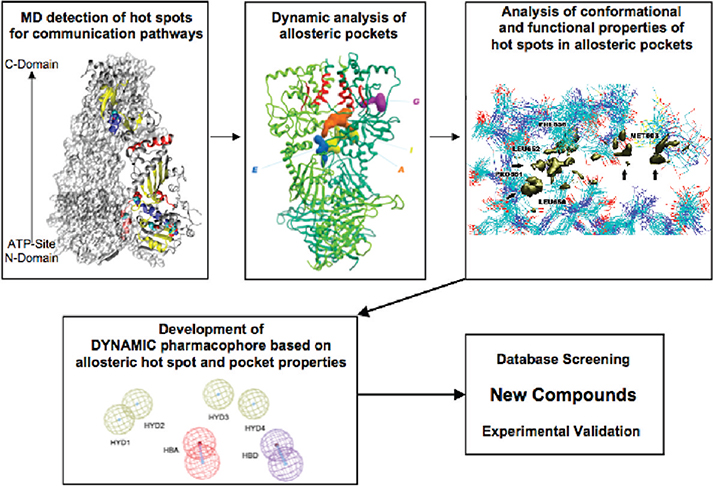
Scheme 1. Computational Biology Strategy for the Selection of Allosteric Inhibitors Exploiting Protein Dynamicsa.
a From the analysis of the dynamics of the activated state of the protein (Hsp90 bound to ATP), the hot spot residues active in mediating signal transduction are identified. Analysis of possible binding pockets centered on these residues identifies putative allosteric binding sites. A consensus model of functional interactions with the hot spots together with structural shape constraints is used for pharmacophore modeling. The ensemble-based approach ensures the incorporation of receptor flexibility into the pharmacophore model. Small molecule databases are then screened for leads fitting with the pharmacophoric hypothesis, and selected hits are tested experimentally.