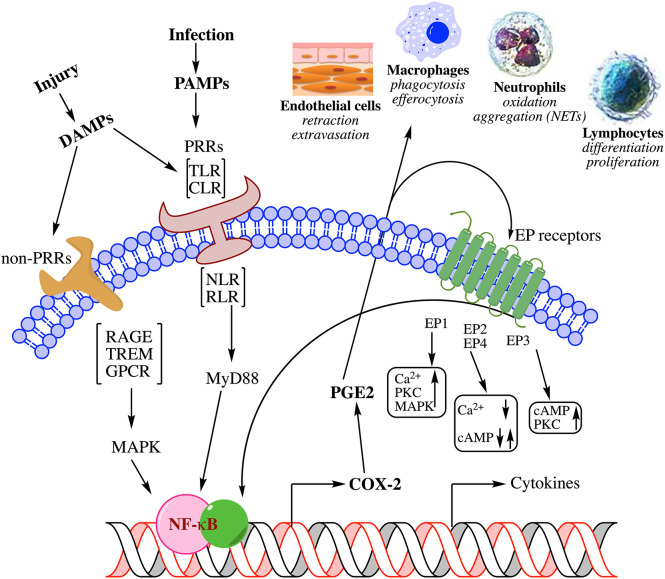
Figure 1.
Summary of biochemical pathways leading to inflammation in response to injury. Physical or pathogen driven injuries are perceived by Pattern Recognition Receptors [PRR; e.g., Toll-Like Receptors (TLR), C-type Lectin Receptors (CLR), Nucleotide-binding Oligomerization Domain (NOD)-Like Receptors (NLR), Retinoic acid-Inducible Gene (RIG)-I-Like Receptor (RLR)] as well as non-PRRs [e.g., Receptor for Activated Glycated End products (RAGE), Triggering Receptors Expressed on Myeloid cells (TREM), G-Protein Coupled Receptors (GPCR)], whose ligands include damaged cell debris (DAMP) of the host or invading pathogen (PAMP). These receptors activate transcription factors such as Nuclear Factor kappa-light-chain-enhancer of activated B cells (NF-κB) via adapter proteins like Myeloid Differentiation 88 (MyD88) and Mitogen-Activated Protein Kinase (MAPK) that induce the biosynthesis of lipid mediators such as prostaglandin E2 (PGE2) through cyclooxygenase-2 (COX-2) induction and release of cytokines. These mediators mutually amplify their response both through autocrine and paracrine activation of specific receptors [receptors (EP) for PGE2 shown here] as well as induce endothelial retraction and extravasation, phagocytosis and efferocytosis by macrophages, oxidative metabolism of pathogens by neutrophils, and induction of immune response by lymphocyte differentiation; all manifest in the clinical symptoms of inflammation. For a detailed review on the biochemistry of inflammation and the role of lipid mediators please see these comprehensive reviews (7–10).