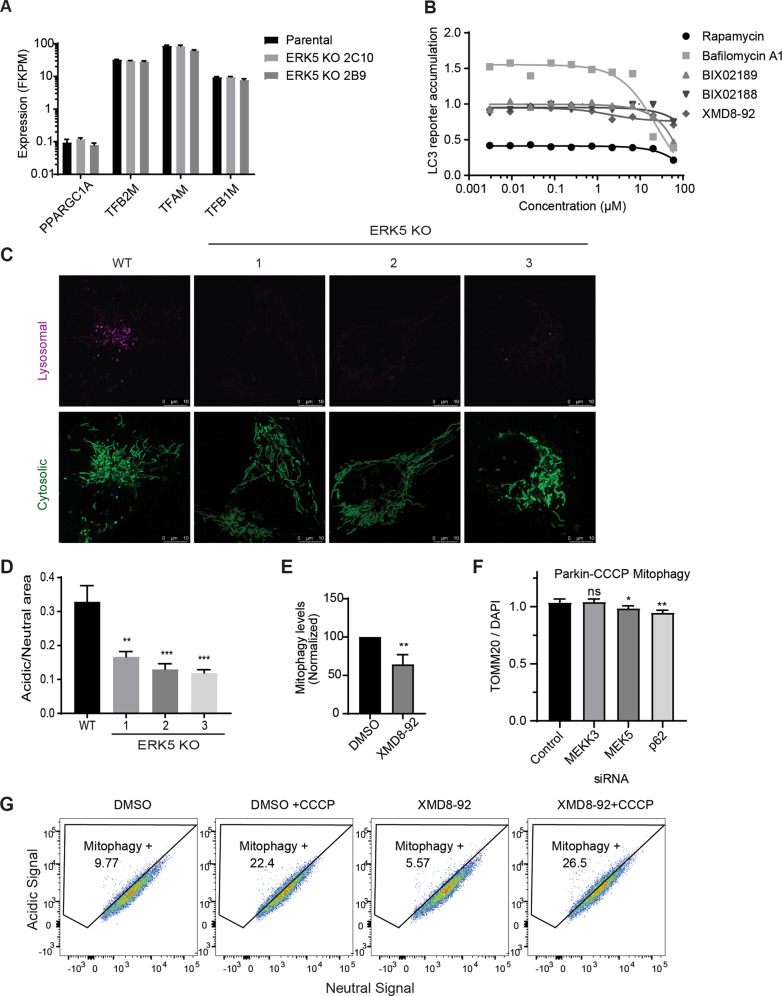
Fig. 4. The MEKK3-MEK5-ERK5 pathway is required for lysosomal degradation of mitochondria.
a RNA-seq was performed on parental U2OS cells and two MAPK7/ERK5-knockout U2OS clones. Expression of several mRNAs encoding mitochondrial biogenesis genes is shown. b HEK293 cells stably expressing the HiBiT-LC3 bulk-autophagy reporter were treated with vehicle (DMSO) or the indicated compounds for 24 h in duplicate. Accumulation of the reporter was measured using the Promega Autophagy Assay system and normalized to vehicle-treated cells. c, d Representative images (c) and quantification (d) of active mitophagy events in parental (WT) and ERK5-knockout U2OS cells expressing mitochondrial-targeted mKeima. e Mitophagy levels in parental U2OS cells expressing mitochondrial-targeted mKeima were assessed by flow cytometry and quantified. Cells were treated overnight with either DMSO or 10 µM XMD8–92. Mean ± SD of n = 3 independent experiments is shown. f HeLa YFP-Parkin stable cells were transfected with the indicated siRNA for 72 h and treated with 10 µM CCCP overnight. Cells were fixed, stained with anti-Tom20 and DAPI, and imaged. Mitochondrial content was quantified and normalized to DAPI. g WT U2OS cells expressing mitochondrial-targeted mKeima were treated overnight with vehicle control, 10 µM CCCP, 10 µM XMD8–92, or both. Mitophagy levels were analyzed via flow cytometry.