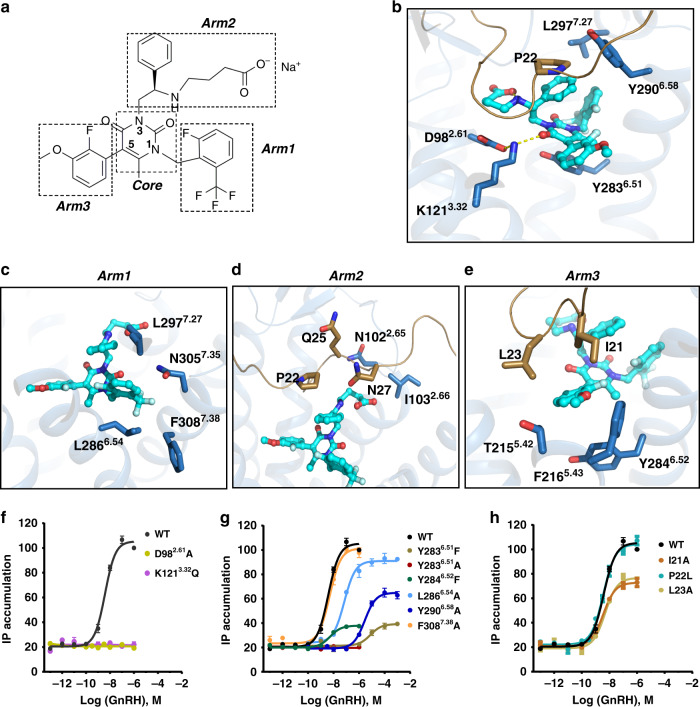
Fig. 2. Interaction of elagolix with GnRH1R.
a Two-dimensional structure of elagolix, the positions 1, 3, and 5 of elagolix uracil core are termed arm1, arm2, and arm3, respectively. b–e Key residues (sky blue sticks) involved in different parts of ligand binding are displayed in b (core), c (arm1), d (arm2), and e (arm3), respectively. The only hydrogen bonds net was shown as dotted yellow lines. f Agonist GnRH dose-dependent responses of GnRH1R wild-type (WT), D982.61A, and K1213.32Q mutants by IP (inositol phosphate) accumulation assays. Both mutation abolished the GnRH-induced signaling. EC50 values are expressed as means ± SEM (n = 3) at three times independently experiment repeats with similar results. g Representative of effects of hydrophobic residues in orthosteric site on agonism of GnRH in IP accumulation assays. EC50 values are expressed as means ± SEM (n = 3) at three times independently experiment repeats with similar results. h Representative of effects of N-terminal residues of the receptor on agonism of GnRH in IP accumulation assays. HEK293T were transiently transfected with the wild-type or mutant receptors (without PGS), and IP accumulation was measured after stimulation with GnRH ligands for 2 h. EC50 values are expressed as means ± SEM (n = 3) at three times independently experiment repeats with similar results.