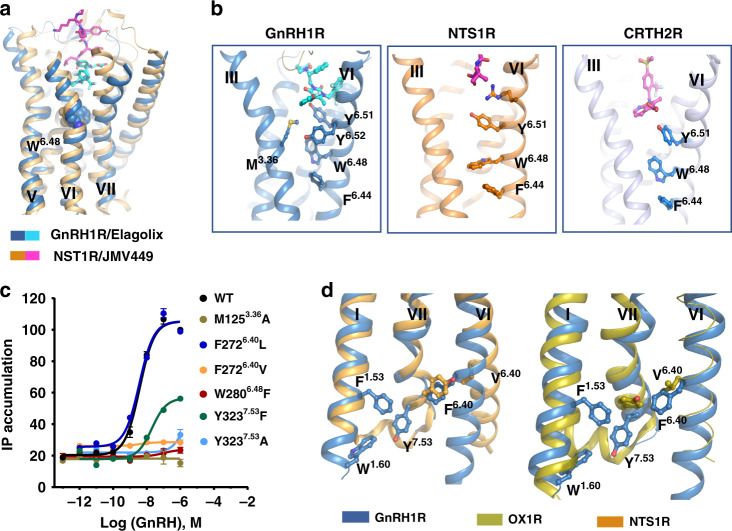
Fig. 5. The structural features of microswitches in GnRH1R.
a Structural superposition of GnRH1R with active NTS1R (PDB ID: 6OS9, orange cartoon). Agonist JMV449 of NTS1R is shown magentas. Both elagolix and JMV449 bound to a shallow pocket that is located near the extracellular region of those two receptors. b The side chains of the hydrophobic Y6.51-W6.48-F6.44 motif in TM6 are shown as sticks in GnRH1R (left), NTS1R (middle, PDB ID: 4S0V), and CRTH2R (right, PDB ID: 6D26), respectively. c Effects of GnRH agonist on activating GnRH1R WT, M1253.36A, F2726.40L, F2726.40V, W2806.48F, Y3237.53F, and Y3237.53A mutants expressed in HEK293T cells independently, monitored by IP1 accumulation assays. HEK293T were transiently transfected with the wild-type or mutant receptors (without PGS) and IP accumulation was measured after stimulation with GnRH ligands for 2 h. EC50 values are expressed as means ± SEM (n = 3) at three times independently experiment repeats with similar results. d The rotamer comparison of NP7.50xxY7.53 motif of GnRH1R with inactive OX2R (PDB ID: 4S0V, green cartoon) and active NTS1R (PDB ID: 6OS9, orange cartoon) reveals an unusual conformation of Y7.53 in GnRH1R. The side chain of F1.53, W1.60, and F6.40 residues are shown as sticks with corresponding color to their receptors.