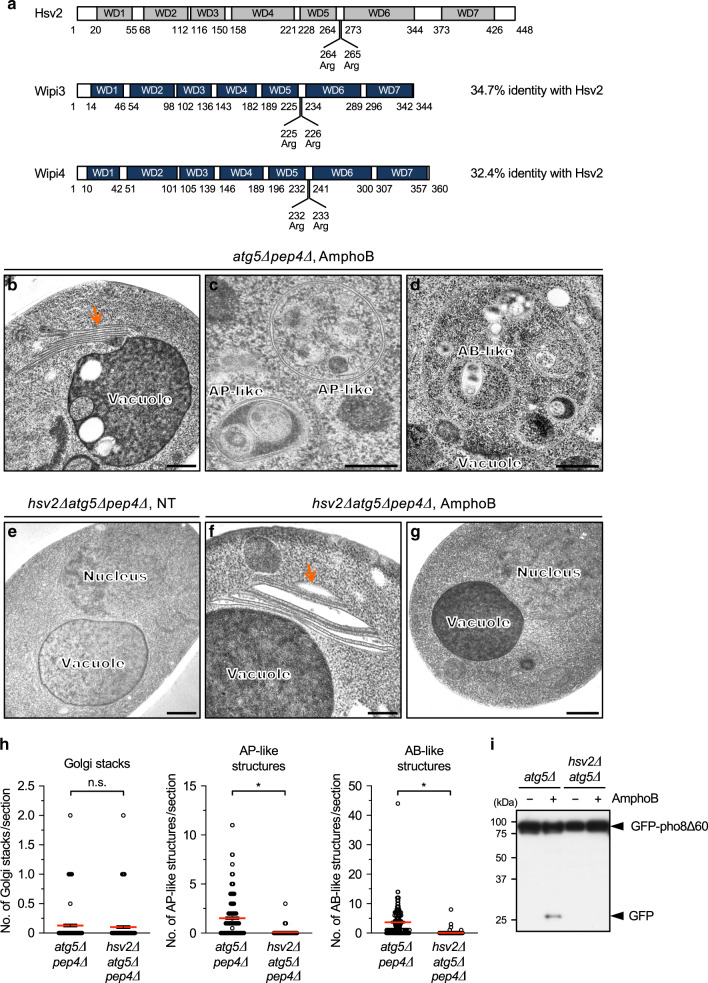
Fig. 1. Hsv2 is a crucial molecule for alternative autophagic responses in yeast.
a Structure of Hsv2 and its murine homologues Wipi3 and Wipi4. Seven WD domains and PIP-interacting sites (two arginine residues) are indicated. Amino acid numbers are shown below each protein. b–g Requirement of Hsv2 for the AmphoB-induced alternative autophagic response. atg5∆pep4∆ yeast cells (b–d) and hsv2∆atg5∆pep4∆ yeast cells (e–g) were not (NT) or were treated with AmphoB (2.5 µg/mL for 24 h) and observed using electron microscopy (EM). In atg5∆pep4∆ cells, Golgi stacking (b, arrow), autophagosome (AP)-like structures (c) and autophagic body (AB)-like structures (d) were observed. In hsv2∆atg5∆pep4∆ cells, stacked and swollen Golgi membranes (arrow) were observed (f). AP-like and AB-like structures were not observed in the cytosol and vacuoles, respectively (g). Bars = 0.5 µm (b, d–g) and 0.2 µm (c). h The number of Golgi stacks, and AP-like and AB-like structures per section are shown as the mean ± SEM (atg5∆pep4∆: n > 191 cells; hsv2∆atg5∆pep4∆: n > 100 cells). Red bars indicate mean values. Comparisons were performed using unpaired two-tailed Student t-tests. Left panel: ns: not significant (p = 0.4137). Other panels: *p < 0.01 (exact p values cannot be described since the value is too small [p < 0.0001]). i GFP–pho8∆60-expressing atg5∆ cells and hsv2∆atg5∆ cells were treated with or without AmphoB (2.5 µg/mL for 24 h), and subjected to western blotting. The generation of free GFP is a marker of the autophagic response. Source data are provided as a Source Data file.