Figure 2.
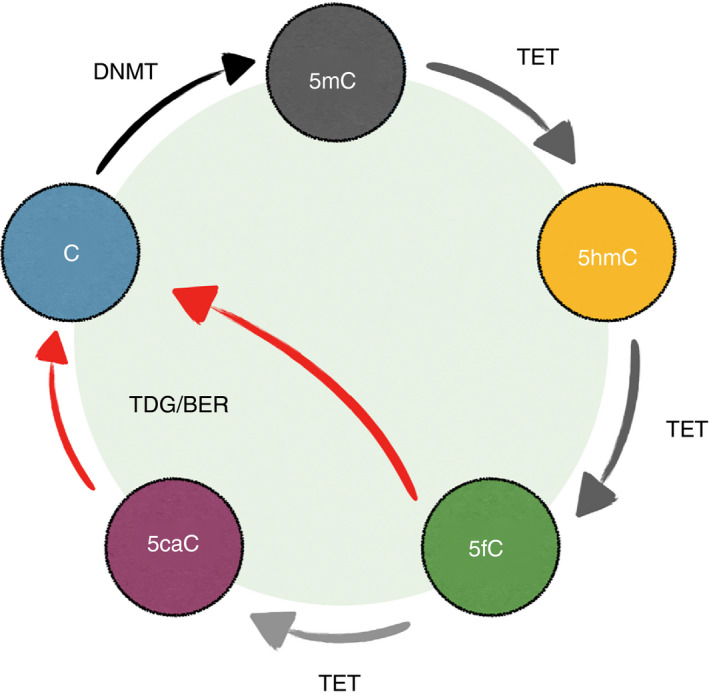
The DNA methylation cycle. In the mammalian genome, the majority of the cytosines at CG motifs are methylated. DNA methyltransferases (DNMTs) catalyze the addition of a methyl group to the fifth carbon of cytosine (C), generating 5‐methylcytosine (5mC). TETs then convert 5mC into oxidized methylcytosines (oxi‐mCs): 5‐hydroxymethylcytosine (5hmC), 5‐formylcytosine (5fC), and 5‐carboxylcytosine (5caC). While TETs are capable of the complete oxidization of 5mC to 5caC in vitro, the majority of oxi‐mCs in the cells are 5hmC. 5hmC is stable and a potential epigenetic mark. 5fC and 5caC are unstable and are removed by TDG (thymine DNA glycosylase) with the base‐excision repair. The base removal process (red arrows) constitutes ‘active DNA demethylation’. During DNA replication, the pairing between newly synthesized DNA with the original modified CpG motif creates the hemi‐modified CpG. The maintenance DNA methyltransferase complex DNMT1/UHRF1 recognizes the hemi‐methylated CpG and methylates the unmodified cytosine on the new DNA. However, DNMT1/UHRF1 cannot recognize the hemi‐methylated CpG containing oxi‐mCs, preventing the methylation of the newly synthesized DNA. Therefore, the methylation pattern will be erased after rounds of DNA replication
