Lomentospora prolificans is an opportunistic fungal pathogen with low susceptibility to current antifungal drugs. Here, we tested the in vitro susceptibility of 8 drugs against 42 clinical L. prolificans isolates. All isolates showed high MICs to voriconazole (MIC90>16 μg/ml), itraconazole (MIC90>16 μg/ml), posaconazole (MIC90>16 μg/ml), isavuconazole (MIC90>16 μg/ml), amphotericin B (MIC90>16 μg/ml), and terbinafine (MIC90>64 μg/ml) and high minimum effective concentrations (MECs) to micafungin (MEC90>8 μg/ml), with the exception of miltefosine showing an MIC90 value of 4 μg/ml.
KEYWORDS: Lomentospora prolificans, antifungal susceptibility, synergy, resistance mechanisms, antifungal agents, antifungal resistance, antifungal susceptibility testing
ABSTRACT
Lomentospora prolificans is an opportunistic fungal pathogen with low susceptibility to current antifungal drugs. Here, we tested the in vitro susceptibility of 8 drugs against 42 clinical L. prolificans isolates. All isolates showed high MICs to voriconazole (MIC90>16 μg/ml), itraconazole (MIC90>16 μg/ml), posaconazole (MIC90>16 μg/ml), isavuconazole (MIC90>16 μg/ml), amphotericin B (MIC90>16 μg/ml), and terbinafine (MIC90>64 μg/ml) and high minimum effective concentrations (MECs) to micafungin (MEC90>8 μg/ml), with the exception of miltefosine showing an MIC90 value of 4 μg/ml. We examined six different in vitro drug combinations and found that the combination of voriconazole and terbinafine achieved the most synergistic effort against L. prolificans. We then annotated the L. prolificans whole genome and located its Cyp51 and Fks1 genes. We completely sequenced the two genes to determine if any mutation would be related to azole and echinocandin resistance in L. prolificans. We found no amino acid changes in Cyp51 protein and no tandem repeats in the 5′ upstream region of the Cyp51 gene. However, we identified three intrinsic amino acid residues (G138S, M220I, and T289A) in the Cyp51 protein that were linked to azole resistance. Likewise, two intrinsic amino acid residues (F639Y, W695F) that have reported to confer echinocandin resistance were found in Fks1 hot spot regions. In addition, three new amino acid alterations (D440A, S634R, and H1245R) were found outside Fks1 hot spot regions, and their contributions to echinocandin resistance need future investigation. Overall, our findings support the notion that L. prolificans is intrinsically resistant to azoles and echinocandins.
INTRODUCTION
Lomentospora prolificans, formerly known as Scedosporium prolificans, is an opportunistic fungal pathogen that causes superficial to invasive fungal infections in immunocompromised and occasionally in immunocompetent people (1–3). L. prolificans has been reported to be resistant to a wide range of antifungal drugs, and its infection is often accompanied by high mortality rates (3, 4). Therefore, new effective antifungal strategies are urgently needed.
Antifungal drug combination therapy may be a useful approach. Several studies displayed that combinations of terbinafine with azoles or micafungin with amphotericin B had achieved a synergistic effect in vitro in some L. prolificans isolates (5–7) as well as demonstrated a therapeutic response in several clinical cases (8–10). Miltefosine, a kind of alkyl-phospholipid analogue, could be another option for treating L. prolificans infection. Although the drug was originally developed as an antileishmanial agent, it was found to possess antifungal activity (11–13). In vitro studies also demonstrated that combinations of miltefosine with antifungal drugs had a synergistic effect against some clinical mold isolates (13–15).
The antifungal drug-resistant mechanism in L. prolificans is poorly understood, and more studies are needed to understand its resistance mechanisms to azoles and echinocandins (16). Amino acid substitutions of the Cyp51 protein and tandem repeats in the promoter region of the Cyp51 gene are the common azole resistance mechanisms in filamentous fungal pathogens (17). A partial protein sequence of Cyp51 in L. prolificans was previously reported, and several intrinsic amino acid residues associated with azole resistance in Aspergillus fumigatus were identified by a conserved residues amino acid alignment (18). Similarly, amino acid substitutions of Fks1 are a common echinocandin-resistant mechanism in other fungal pathogens (19). By hybrid expression of the partial Fks1 sequence of L. prolificans in Saccharomyces cerevisiae, it was demonstrated that the innate or intrinsic amino acid residues in L. prolificans Fks1 equivalent to S. cerevisiae W695F and F639Y mutations in Fks1 hot spot regions may be linked to the intrinsic echinocandin resistance in L. prolificans (20, 21). However, both the complete open reading frames of Cyp51 and Fks1 genes are not revealed, and no data are shown about the gene polymorphisms of Cyp51 and Fks1 in clinical L. prolificans isolates.
In this study, we tested the antifungal susceptibility of 42 clinical isolates, including testing the efficacy of various in vitro drug combinations. In addition, we sequenced both complete open reading frames of Cyp51 and Fks1 genes and analyzed the azole and echinocandin resistance mechanisms in clinical L. prolificans isolates.
RESULTS
Clinical information of isolates and their antifungal susceptibility.
A total of 42 clinical isolates was collected from 13 patients (Table 1). Seven patients had multiple isolates recovered from different dates. Most isolates were isolated from pulmonary sources, followed by two isolates from blood, one isolate from a cornea, and one from a wound. As for antifungal susceptibility profiles, all isolates except one isolate (ZX2) were found to have the highest MICs for azoles and terbinafine (Table 1). For itraconazole, voriconazole, posaconazole, and isavuconazole, the values of median MIC, mode MIC, MIC50, and MIC90 were all over 16 μg/ml, and values of the geometric mean MIC were 30.97, 27.58, 30.45, and 28.51 μg/ml, respectively. Although micafungin and amphotericin B showed little activity against some clinical isolates (Table 1), their values of median MIC, mode MIC, minimum effective concentration at which 50% of the isolates tested are effective (MEC50) or MIC50, and MEC90 or MIC90 were all >8 and >16 μg/ml, respectively, and their geometric mean MIC values were 8.98 and 20.84 μg/ml, respectively. Terbinafine had the highest MIC values with median MIC, mode MIC, MIC50, and MIC90 all >64 μg/ml and geometric mean MIC of 125.90 μl/ml. Miltefosine was the only drug that showed the lowest MICs against all L. prolificans isolates, with median MIC, mode MIC, MIC50, and MIC90 values all of 4 μg/ml and geometric mean MIC of 3.12 μg/ml, suggesting that miltefosine may possess some good activity against L. prolificans compared to these antifungal drugs.
TABLE 1.
Clinical information of isolates and their drug susceptibility profiles
| Pa | Disease | IDb | Date (yr/mo/day)c | Source | MIC (μg/ml) of: |
MICA MEC (μg/ml) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AMB | ITC | VOR | POS | ISA | TB | MIL | ||||||
| P1 | CF | 117d ,e | 2014/8/17 | S | >16 | >16 | >16 | >16 | >16 | >64 | 4 | >8 |
| 119d | 2014/8/21 | S | >16 | >16 | >16 | >16 | >16 | >64 | 4 | >8 | ||
| 120 | 2014/8/21 | S | >16 | >16 | >16 | >16 | >16 | >64 | 4 | 8 | ||
| 126e | 2014/9/11 | S | >16 | >16 | >16 | >16 | >16 | >64 | 4 | 2 | ||
| 128d ,e | 2014/9/11 | S | >16 | >16 | >16 | >16 | >16 | >64 | 2 | >8 | ||
| 143d ,e | 2014/12/23 | S | >16 | >16 | >16 | >16 | >16 | >64 | 2 | >8 | ||
| ZX1e | 2015/1/20 | S | >16 | >16 | >16 | >16 | >16 | >64 | 4 | 2 | ||
| 144d ,e | 2015/3/17 | S | >16 | >16 | >16 | >16 | >16 | >64 | 4 | >8 | ||
| 163 | 2015/7/13 | BAL | >16 | >16 | >16 | >16 | >16 | >64 | 4 | 8 | ||
| 166d ,e | 2015/8/18 | S | >16 | >16 | >16 | >16 | >16 | >64 | 2 | >8 | ||
| 179e | 2015/12/17 | S | 16 | >16 | >16 | >16 | >16 | >64 | 4 | 4 | ||
| 181d ,e | 2015/12/17 | S | >16 | >16 | >16 | >16 | >16 | >64 | 2 | >8 | ||
| 182d | 2015/12/17 | S | >16 | >16 | >16 | >16 | >16 | >64 | 2 | >8 | ||
| P2 | CF | 17 | 2012/6/8 | S | 4 | >16 | 16 | >16 | >16 | >64 | 4 | >8 |
| 19 | 2012/6/8 | S | >16 | >16 | >16 | >16 | >16 | >64 | 4 | 8 | ||
| 27 | 2012/6/27 | S | >16 | >16 | 16 | >16 | >16 | >64 | 4 | >8 | ||
| 35d | 2012/7/25 | T | >16 | >16 | 16 | >16 | 16 | >64 | 2 | >8 | ||
| 72d ,e | 2013/1/5 | E | >16 | >16 | >16 | >16 | >16 | >64 | 2 | >8 | ||
| P3 | CF | 23d | 2012/5/31 | BAL | >16 | >16 | >16 | >16 | >16 | >64 | 4 | >8 |
| 24e | 2012/6/13 | BAL | 8 | >16 | >16 | >16 | 16 | >64 | 2 | 2 | ||
| 22e | 2012/6/25 | BAL | 4 | >16 | 16 | >16 | 16 | >64 | 2 | >8 | ||
| 29e | 2012/6/25 | BAL | >16 | >16 | >16 | >16 | >16 | >64 | 4 | 4 | ||
| 25 | 2012/7/2 | S | 4 | >16 | 16 | >16 | >16 | >64 | 4 | >8 | ||
| P4 | CF | ZX2e | 2014/10/1 | S | 16 | 8 | 4 | 4 | 4 | 64 | 2 | 2 |
| 145e | 2015/3/6 | BAL | 8 | >16 | 16 | >16 | 16 | >64 | 4 | 1 | ||
| ZX3 | 2018/1/5 | S | 2 | >16 | >16 | >16 | >16 | >64 | 1 | >8 | ||
| ZX4 | 2018/4/20 | S | >16 | >16 | >16 | >16 | >16 | >64 | 4 | >8 | ||
| ZX5 | 2018/4/20 | S | 16 | >16 | >16 | >16 | >16 | >64 | 4 | 8 | ||
| ZX6e | 2018/4/20 | S | 4 | >16 | >16 | >16 | >16 | >64 | 2 | >8 | ||
| P5 | MC | ZX8 | 2018/9/5 | Tb | >16 | >16 | >16 | >16 | >16 | >64 | 4 | 8 |
| ZX9e | 2018/9/5 | Tb | >16 | >16 | >16 | >16 | >16 | >64 | 4 | 4 | ||
| ZX10 | 2018/9/5 | BAL | >16 | >16 | >16 | >16 | >16 | >64 | 4 | 8 | ||
| P6 | AML | ZX11d ,e | 2018/5/22 | Blood | >16 | >16 | >16 | >16 | >16 | >64 | 4 | >8 |
| ZX12d | 2018/5/22 | Blood | >16 | >16 | >16 | >16 | >16 | >64 | 4 | >8 | ||
| P7 | CF | 216 | 2016/11/3 | T | >16 | >16 | >16 | >16 | >16 | >64 | 4 | 8 |
| 220 | 2017/2/9 | S | >16 | >16 | >16 | >16 | >16 | >64 | 4 | 8 | ||
| P8 | CF | ZX16 | 2019/4/9 | S | 4 | >16 | >16 | >16 | >16 | >64 | 4 | 8 |
| P9 | CU | ZX13d ,e | 2014/2/11 | Cornea | >16 | >16 | >16 | >16 | >16 | >64 | 2 | >8 |
| P10 | AIDS | 84 | 2013/12/16 | S | >16 | >16 | >16 | >16 | >16 | >64 | 4 | 8 |
| P11 | CS | 176d ,e | 2015/12/2 | E | >16 | >16 | >16 | >16 | >16 | >64 | 2 | >8 |
| P12 | N | ZX14 | 2015/5/7 | Wound | >16 | >16 | >16 | >16 | >16 | >64 | 4 | 8 |
| P13 | SOD | ZX15e | 2018/7/8 | E | >16 | >16 | >16 | >16 | >16 | >64 | 4 | 4 |
P, patient; AMB, amphotericin B; ITC, itraconazole; VOR, voriconazole; POS, posaconazole; ISA, isavuconazole; TB, terbinafine; MIL, miltefosine; MICA, micafungin; CF, cystic fibrosis; MC, multiple cancers; AML, acute myeloblastic leukemia; CU, corneal ulcer; CS, chronic sinusitis; N, not available; SOD, solid organ transplantation; S, sputum (expectorated); T, throat swabs; E, endotrachial aspirate; BAL fluid, bronchoalveolar lavage fluid; Tb, transbronchial biopsy.
ID, isolate number.
Sample collection date.
Isolates (a total of 15 isolates) were tested in vitro susceptibility for different antifungal drug combinations shown in Table 2 to 4.
Isolates (a total of 21 isolates) were sequenced for Cyp51 and FKs1 genes.
Antifungal synergy testing.
Fifteen L. prolificans isolates were tested in vitro for drug susceptibility for different drug combinations (Tables 2 to 4). A synergistic effect of terbinafine-voriconazole was observed in seven clinical isolates (47%). Notably, the MIC of terbinafine was reduced over 32-fold (from >64 to 2 μg/ml) in isolate 176, and the MIC of voriconazole was reduced 4-fold (16 to 4 μg/ml) in isolate 35 (Table 2). However, the synergy effect of the terbinafine-amphotericin B combination was only observed in one isolate (isolate 23) (Table 2). When the voriconazole-amphotericin B combination was tested, only two clinical isolates demonstrated a synergistic effect (isolates 117 and 23); no synergistic effect was observed in the voriconazole-micafungin combination (Table 3). When the amphotericin B-miltefosine combination was tested, the amphotericin B MIC decreased at least 64-fold (from >16 to 0.25 μg/ml) in three isolates (117, 119, and ZX12), but a synergistic effect was only observed in two isolates (isolate 23 and ZX11); no synergistic effect was observed in the voriconazole-miltefosine combination (Table 4).
TABLE 2.
Synergy test results of AMB, TB, and VOR against L. prolificans based on FICI values
| Isolate | MIC (μg/ml) of: |
FICIa | MIC (μg/ml) of: |
FICIa | ||||
|---|---|---|---|---|---|---|---|---|
| AMBb | TB | AMB/TB | VOR | TB | VOR/TB | |||
| 35 | >16 | >64 | >16/>64 | 2 | 16 | >64 | 4/32 | 0.5 |
| 117 | >16 | >64 | >16/>64 | 2 | >16 | >64 | 8/8 | 0.317 |
| 119 | >16 | >64 | >16/>64 | 2 | >16 | >64 | 8/32 | 0.5 |
| ZX11 | >16 | >64 | >16/>64 | 2 | >16 | >64 | 16/16 | 0.625 |
| 23 | >16 | >64 | 4/8 | 0.281 | >16 | >64 | 8/32 | 0.5 |
| ZX12 | >16 | >64 | >16/>64 | 2 | >16 | >64 | 16/8 | 0.563 |
| 143 | >16 | >64 | >16/>64 | 2 | >16 | >64 | 16/32 | 0.75 |
| 128 | >16 | >64 | >16/>64 | 2 | >16 | >64 | 8/8 | 0.313 |
| 182 | >16 | >64 | >16/>64 | 2 | >16 | >64 | >16/>64 | 2 |
| 181 | >16 | >64 | >16/>64 | 2 | >16 | >64 | >16/>64 | 2 |
| 166 | >16 | >64 | >16/>64 | 2 | >16 | >64 | >16/>64 | 2 |
| 144 | >16 | >64 | >16/>64 | 2 | >16 | >64 | 16/32 | 0.75 |
| 72 | >16 | >64 | >16/>64 | 2 | >16 | >64 | 16/0.5 | 0.504 |
| ZX13 | >16 | >64 | >16/>64 | 2 | >16 | >64 | 8/32 | 0.5 |
| 176 | >16 | >64 | >16/>64 | 2 | >16 | >64 | 8/2 | 0.266 |
FICI values in bold indicate synergy.
AMB, amphotericin B; TB, terbinafine; VOR, voriconazole.
TABLE 3.
Synergy test results of AMB, VOR, and MICA against L. prolificans based on FICI values
| Isolate | MIC (μg/ml) of: |
FICIa | MIC/MEC (μg/ml) of: |
FICIa | ||||
|---|---|---|---|---|---|---|---|---|
| AMBc | VOR | AMB/VOR | VOR | MICA | VOR/MICAb | |||
| 35 | >16 | 16 | 16/4 | 0.75 | 16 | >16 | 16/8 | 1.25 |
| 117 | >16 | >16 | 8/4 | 0.375 | >16 | 16 | >16/>16 | 3 |
| 119 | >16 | >16 | 1/16 | 0.531 | >16 | >16 | >16/>16 | 2 |
| ZX11 | >16 | >16 | 4/16 | 0.625 | >16 | >16 | >16/>16 | 2 |
| 23 | >16 | >16 | 1/8 | 0.281 | >16 | 16 | 16/8 | 1 |
| ZX12 | >16 | >16 | 4/16 | 0.625 | >16 | >16 | >16/>16 | 2 |
| 143 | >16 | >16 | 8/16 | 0.75 | >16 | >16 | >16/>16 | 2 |
| 128 | >16 | >16 | 2/16 | 0.562 | >16 | 16 | >16/>16 | 3 |
| 182 | >16 | >16 | >16/>16 | 2 | >16 | >16 | >16/>16 | 2 |
| 181 | >16 | >16 | >16/>16 | 2 | >16 | >16 | >16/>16 | 2 |
| 166 | >16 | >16 | >16/>16 | 2 | >16 | >16 | >16/>16 | 2 |
| 144 | >16 | >16 | >16/>16 | 2 | >16 | 16 | 16/16 | 1.5 |
| 72 | >16 | >16 | >16/>16 | 2 | >16 | >16 | >16/>16 | 2 |
| ZX13 | >16 | >16 | >16/>16 | 2 | >16 | >16 | >16/>16 | 2 |
| 176 | >16 | >16 | >16/>16 | 2 | >16 | >16 | >16/>16 | 2 |
FICI values in bold indicate synergy.
MICs obtained in combination in checkerboard assays.
AMB, amphotericin B; VOR, voriconazole; MICA, micafungin.
TABLE 4.
Synergy test results of AMB, MIL, and VOR against L. prolificans based on FICI values
| Isolate | MIC (μg/ml) of: |
FICIa | MIC (μg/ml) of: |
FICIa | ||||
|---|---|---|---|---|---|---|---|---|
| AMBb | MIL | AMB/MIL | VOR | MIL | VOR/MIL | |||
| 35 | >16 | 2 | >16/>1 | 2 | 16 | 2 | 16/>1 | 2 |
| 117 | >16 | 4 | 0.25/2 | 0.508 | >16 | 4 | 16/2 | 1 |
| 119 | >16 | 4 | 0.25/2 | 0.508 | >16 | 4 | 16/2 | 1 |
| ZX11 | >16 | 4 | 8/0.5 | 0.375 | >16 | 4 | 16/2 | 1 |
| 23 | >16 | 4 | 8/0.5 | 0.375 | >16 | 4 | 16/0.06 | 0.515 |
| ZX12 | >16 | 4 | 0.25/2 | 0.508 | >16 | 4 | 16/2 | 1 |
| 143 | >16 | 2 | 4/1 | 0.625 | >16 | 2 | >16/>1 | 2 |
| 128 | >16 | 2 | >16/>1 | 2 | >16 | 2 | 16/1 | 1 |
| 182 | >16 | 2 | >16/>1 | 2 | >16 | 2 | >16/>1 | 2 |
| 181 | >16 | 2 | >16/>1 | 2 | >16 | 2 | >16/>1 | 2 |
| 166 | >16 | 2 | >16/>1 | 2 | >16 | 2 | >16/>1 | 2 |
| 144 | >16 | 4 | 0.5/2 | 0.516 | >16 | 4 | 16/2 | 1 |
| 72 | >16 | 2 | >16/>1 | 2 | >16 | 2 | 16/0.5 | 0.75 |
| ZX13 | >16 | 2 | 16/1 | 1 | >16 | 2 | >16/>1 | 2 |
| 176 | >16 | 2 | >16/>1 | 2 | >16 | 2 | >16/>1 | 2 |
FICI values in bold indicate synergy.
AMB, amphotericin B; MIL, miltefosine; VOR, voriconazole.
Sequence analysis of the Cyp51 gene and promoter region in clinical isolates.
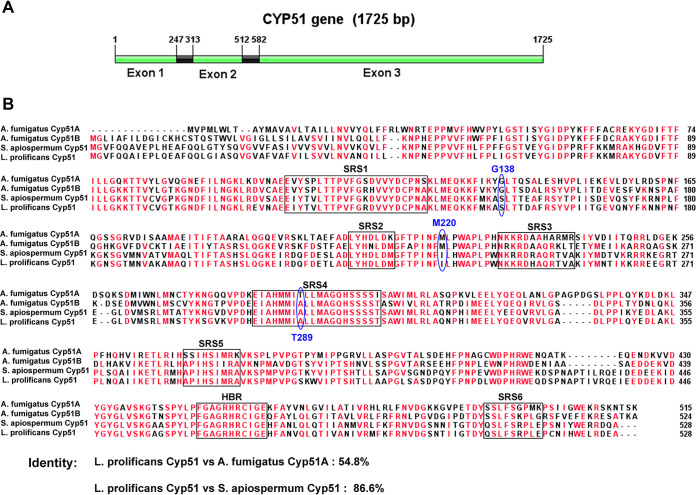
The complete open reading frame of the L. prolificans Cyp51 gene was determined by bioinformatics analysis and showed that the Cyp51 gene has 1,725 bp and 2 predicted exons (Fig. 1A). In order to identify amino acid substitutions in the Cyp51 protein that could be associated with azole resistance, the Cyp51 gene was sequenced in 21 L. prolificans clinical isolates (Table 5). None of these isolates displayed any amino acid substitutions in the Cyp51 protein compared to the ones that are sequence-annotated from the L. prolificans whole-genome sequencing (GenPept accession no. PKS10573.1). However, the synonymous mutation T1530A was found in two isolates. The isolate ZX2 showed relatively the lowest MICs of azoles compared to other isolates, yet its Cyp51 protein sequence did not differ from that of the others isolates with high MICs to azoles. By aligning the amino acid sequence of Cyp51 from A. fumigatus, Scedosporium apiospermum, and L. prolificans, we found the identity between Cyp51 of S. apiospermum and Cyp51 of L. prolificans was 86.6% (Fig. 1B). Additionally, regions of substrate recognition sites (SRSs) of P450 and the heme-binding region (HBR) among these species were conserved. Notably, the L. prolificans Cyp51 protein contained residues at three locations (S153, I235, and A302) that corresponded to three residues (G138, M220, and T289) in A. fumigatus Cyp51A (Fig. 1B) whose alterations (G138S, M220I, and T289A) have been reported to be associated with azole resistance (22–24). These innate or intrinsic amino acid residues presented in L. prolificans at the above locations corresponded to respective mutations found in azole-resistant A. fumigatus Cyp51A. In addition, we sequenced the promoter region (1,950 nucleotides upstream of the start codon) of Cyp51 in 21 L. prolificans isolates. We did not identify any tandem repeats in the promoter region, although 4 single nucleotide polymorphisms (SNPs) (C-164G, C-174A, G-1525A, and A-1888G) were found (Table 5).
FIG 1.
(A) Schematic map of the Cyp51 gene open reading frame of L. prolificans. (B) Amino acid sequence alignment of Cyp51 from A. fumigatus, Scedosporium apiospermum, and L. prolificans. The Cyp51 protein sequences shown here originated from NCBI GenBank database (accession nos. XP_752137.1, XP_749134.1, MH120909.1, and MN329109). Residues with the similarity of 75% and above are in red. The conserved substrate recognition sites (SRSs) of P450 and heme-binding region (HBR) are encircled with black boxes. Three amino acid residues whose alterations had been linked to azole resistance in A. fumigatus are circled with blue oval frames. A. fumigatus Cyp51 amino acid numbering is used in this picture.
TABLE 5.
Variability of Cyp51 and Fks1 genes in clinical L. prolificans isolates
| Gene or region | Characteristics of DNA sequences |
Characteristics of protein sequences |
|||
|---|---|---|---|---|---|
| Length (nt) | No. of SNPs | No. of allele types | Amino acid changesa | No. of protein types | |
| Cyp51 | 1,725 | 1 | 2 | 1 | |
| 5′ UTRb | 1,950 | 4 | 5 | ||
| Fks1 | 5,841 | 9 | 8 | D440A, S634R, H1245P | 4 |
5′ UTR, 5′ untranslated region of the Cyp51 gene (promoter region).
Amino acid changes compared with the protein sequence with the highest frequency of prevalence.
Sequence analysis of the FKS1 gene in clinical isolates.
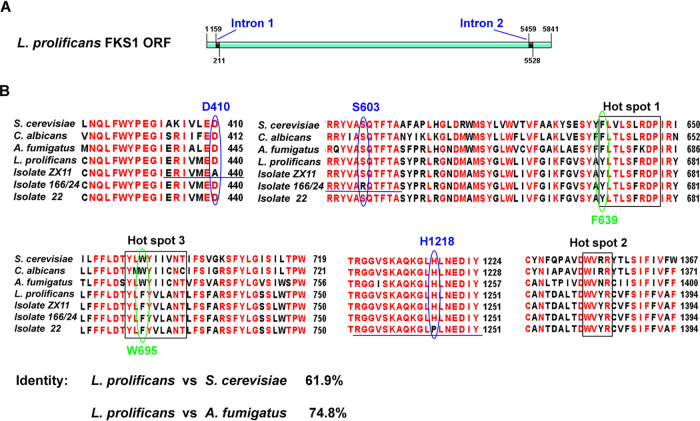
The complete open reading frame of L. prolificans Fks1 gene was determined by bioinformatics analysis and showed that the Fks1 gene has 5,841 bp and 3 predicted exons (Fig. 2A). In order to identify amino acid substitutions in the Fks1 protein that could be associated with echinocandin resistance, the Fks1 gene was sequenced in the 21 L. prolificans clinical isolates. We found 9 SNPs (T164G, A1372C, C1496T, C1955T, A3787C, C3788T, C4202T, C5124T, and C5769A) (Table 5). The SNP T164G in the intron 1 region was found in an isolate with a high micafungin MEC (>8 μg/ml). By aligning amino acid sequences of Fks1 from S. cerevisiae, Candida albicans, A. fumigatus, and L. prolificans (the identity between Fks1 of L. prolificans and Fks1 of S. cerevisiae was 61.9%), we found 3 resulting amino acid substitutions (D440A, S634R, and H1245P) of the Fks1 protein in the conserved regions (Fig. 2B; Table 5) (S. cerevisiae Fks1 amino acid numbering is used). Amino acid substitution D440A was found in one isolate (isolate ZX11) with a micafungin MEC of >8 μg/ml; substitution H1245P was found in a separate isolate (isolate 22) with a micafungin MEC of >8 μg/ml. Additionally, an amino acid substitution S634R was found in one isolate (isolate 24) with an MEC of 2 μg/ml and one isolate (isolate 166) with a micafungin MEC of >8 μg/ml. Notably, all 21 isolates contained two intrinsic amino acid residues in two hot spot regions of Fks1 proteins, F639Y in hot spot 1 and W695F in hot spot 3 (Fig. 2B). Substitutions to these residues have previously been demonstrated to cause echinocandin resistance in other fungi (20, 21).
FIG 2.
(A) Schematic map of Fks1 gene open reading frame of L. prolificans. (B) Amino acid sequence alignment of Fks1 from S. cerevisiae, C. albicans, A. fumigatus, and L. prolificans. The Fks1 protein sequences shown here originated from NCBI GenBank database and our sequenced isolates (accession nos. NC_001144.5, XP_721429.2, XP_751118.1, and MN329116). Residues with the similarity of 75% and above are in red. Three hot spot regions of Fks1 in which amino acid changes introduce echinocandin resistance are encircled with black boxes. Two amino acid residues whose alterations had been linked to echinocandin resistance in L. prolificans are circled with green oval frames. The amino acid residues circled with blue oval frames were equivalent substations (D440A, S634R, and H1245P) found in a few of our clinical isolates. S. cerevisiae Fks1 amino acid numbering is used in this picture.
DISCUSSION
In this study, all L. prolificans clinical isolates were shown to be resistant to all antifungal drugs. Miltefosine may possess good activity against L. prolificans. Although the drug has been described to act against leishmania by inhibiting the biosynthesis of phospholipids, affecting alkyl-lipid metabolism, inducing an apoptosis-like cell death, and disrupting the parasite’s calcium homeostasis (25–27), the exact mechanism of action of miltefosine against fungi is not well understood. Miltefosine likely has different antifungal mechanisms of action from azoles, echinocandins, and polyenes, and it could be a potential drug of choice to treat L. prolificans infections.
Consistent with a previous study (5), the combination of terbinafine and voriconazole has demonstrated the most synergistic effect against L. prolificans in this study. This drug combination regimen has been reported in successfully treating L. prolificans-infected cases (8, 10, 28). A minimal synergistic effect was observed in the drug combination of amphotericin B with voriconazole or miltefosine. The combination of terbinafine and amphotericin B was also tested, but we did not find any good activity against L. prolificans isolates. The azole-echinocandin combination against L. prolificans has not been well explored previously, although this combination displayed a synergistic effect against Aspergillus species (29, 30). Under this combination, we found voriconazole at a high concentration could cause abnormal hyphae growth of L. prolificans strains (data not shown), similar to what was described previously (31); thus, it is not appropriate to use MECs for evaluating the synergistic effect of the combination of voriconazole and micafungin, and we chose to read the MIC endpoint instead of MEC endpoint for this combination. However, we did not find any synergistic effect of this combination. Likewise, we did not find synergistic activity in the combination of miltefosine and voriconazole in this study, consistent with previous studies (14, 15).
Amino acid alterations in Cyp51 or Erg11 in fungi are a commonly occurring mechanism of azole resistance. Although we did not find any amino acid alterations of Cyp51 in our L. prolificans clinical isolates, we identified several azole resistance-related innate or intrinsic amino acid residues in Cyp51 of L. prolificans. Alteration to these residues has been reported to be associated with azole resistance in A. fumigatus (18). Amino acid substitutions G138S, M220I, and T289A of Cyp51A have been reported to be associated with azole resistance in A. fumigatus, with T289A/Y121F/TR46 being the most common azole resistance-related genotype (22, 32). The T440A mutation was noted in our study. Although such a mutation associated with azole resistance in Aspergillus was suggested previously (33), there was no molecular data to support this notion. While the G138C was previously indicated (34), the association of G138S with azole resistance in A. fumigatus was recently reported (22). M220 substitutions conferred high itraconazole MIC and elevated MICs for the other azoles in A. fumigatus (24). Although these substitutions each alone in Cyp51 are not enough to confer azole resistance in Aspergillus (23, 35) or their substitutions could represent a silent mutation (36, 37), the simultaneous presence of three azole resistance-related innate or intrinsic amino acid residues (G138S, M220I, and T289A) in Cyp51 of clinical L. prolificans isolates may support the notion that L. prolificans is intrinsically resistant to azoles. It happened that Cyp51 protein sequence of L. prolificans contained these intrinsic amino acid residues at the respective positions in Cyp51 of azole-resistant A. fumigatus, suggesting that reduced azole susceptibility of L. prolificans may result from the presence of these intrinsic amino acid residues of Cyp51 protein, leading to a reduced affinity to azoles. However, mutations of Cyp51 are not the sole mechanism accounting for azole resistance in Aspergillus. Azole resistance without Cyp51 mutations has been reported in clinical Aspergillus isolates (38–41). Therefore, the intrinsic azole resistance in L. prolificans could be associated with mechanisms other than Cyp51, and that probably needs further study.
Echinocandin resistance commonly involves amino acid mutations in Fks1 hot spot regions during antifungal therapy (19). Innate or intrinsic residues in L. prolificans Fks1 equivalent to S. cerevisiae W695F and F639Y alterations in hot spot regions were confirmed to contribute to echinocandin resistance in S. cerevisiae (20, 21). All 21 isolates in this study possessed these amino acid residues (W695F, F638Y) in hot spot regions (Fig. 2B), possibly playing a role in causing high MECs (MEC90 > 8 μg/ml) to micafungin in these L. prolificans isolates. Other amino acid substitutions in Fks1, such as D440A and H1245R found in isolates with high micafungin MECs, may also play a role in echinocandin resistance, but this will need to be further studied. Nevertheless, echinocandin resistance in L. prolificans could be due to mechanisms other than the Fks1 mutation, since echinocandin resistance in A. fumigatus has been demonstrated by non-Fks1-related mechanisms (42, 43).
One limitation of our study was the lack of genotyping data of these isolates collected from the same patient to determine if these were the same strain. When we chose these isolates from the same patient (Table 1), we considered them as different strains based on the following criteria: (i) collected at different time points (longitudinal collection), (ii) collected from a different source (bronchoalveolar lavage [BAL] fluid, sputum, tissues, etc.), and (iii) presented with different colony morphologies if collected at the same time. These criteria are commonly applied in clinical microbiology laboratories to label these isolates as different strains, and they often trigger antimicrobial susceptibility testing for each strain to determine if different susceptibility profiling presence would impact patient treatment. In fact, some of our strains collected from the same patient did present with different antifungal drug susceptibility profiles (Table 1). For example, isolate 22 and isolate 24 from patient 3 not only showed different antifungal susceptibility profiles but also demonstrated different amino acid substitutions in FKS1. Therefore, it would be reasonable to consider that these strains were different. However, genotyping would be a definitive way to confirm if they were the same strain or different clades, but this approach would not be feasibly accomplished in the manuscript since such a genotyping method for L. prolificans has not been developed and reported in the literature.
In summary, our study demonstrated that none of the antifungal drugs tested in vitro showed any activity against L. prolificans. However, some synergistic activity was observed in drug combinations, with the combination of voriconazole and terbinafine showing the most synergistic effect. Miltefosine, an antileishmania drug, may possess activity against this fungal pathogen. Studies on Cyp51 and Fsk1, the two genes whose mutations are associated with resistance in azole and echinocandin, respectively, revealed that L. prolificans contained innate or intrinsic amino acid residues in which substitutions to these residues at the respective positions have been reported to cause azole and echinocandin resistance in other fungi, supporting the notion that L. prolificans is intrinsically resistant to azoles and echinocandins.
MATERIALS AND METHODS
Isolate identification and growth.
A total of 42 L. prolificans clinical isolates were isolated from 13 patients at the Johns Hopkins Hospital from May 2012 to April 2019. Multiple isolates from the same patient were collected using the following criteria: (i) collected at different time points, (ii) collected from different sources, and (iii) different colony morphologies if collected at the same time (Table 1). Strain typing was not performed in these isolates. All isolates were confirmed as L. prolificans based on their macroscopic and microscopic morphologies, matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) (Bruker biotype), and DNA sequencing targeting ITS and D1D2 regions. All isolates were grown on Sabouraud and potato flake agar plates (PFA) (BD, Sparks, MD) at 30°C to achieve adequate sporulation before antifungal susceptibility testing.
Antifungal susceptibility testing.
The drug MICs or the minimum effective concentrations (MECs) of the 42 clinical isolates were determined using the broth microdilution method recommended by the Clinical and Laboratory Standards Institute M38-A3 and M61 guidelines. The MIC was determined as the lowest concentration producing 100% growth inhibition and was used for all drugs tested except for micafungin. The MEC was determined as the lowest concentration that causes a hyphal structural change from confluent to granular appearance and was used for micafungin. All isolates were grown in pure culture on PFA slants until sufficient conidia were present to prepare an inoculum. Conidial suspensions were incubated at 35°C, and results were read at 72 h for all drugs. Quality control was performed for each drug set every time using the following ATCC strains: Paecilomyces variotii MYA-3630, Candida parapsilosis 22019, and Candida krusei 6258. For MIC and MEC geometric mean calculations, concentrations >16 μg/ml were set to 32 μg/ml, and concentrations >8 μg/ml were set to 16 μg/ml.
Antifungal synergism testing.
Fifteen L. prolificans isolates with the highest MICs for azoles and echinocandins were tested with in vitro antifungal synergy testing for different drug combinations. Testing for antifungal drug interactions was performed in accordance with the broth microdilution checkerboard method. The final concentration of antifungal drugs ranged from 0.015 to 16 μg/ml for voriconazole, amphotericin B, micafungin, and miltefosine and 0.06 to 64 μg/ml for terbinafine. MIC and MEC readings were carried out at 72 h of incubation. For all drug combination groups, the 100% inhibition endpoint was used for the combination of two drugs, including for the combination of voriconazole and micafungin. The MEC endpoint was not chosen for reading the voriconazole and micafungin combination because abnormal hyphae growth was observed in L. prolificans under a high concentration of voriconazole, as described previously (31) and as well as from our own experiences. To assess the in vitro interactions between antifungal drugs, the fractional inhibitory concentration index (FICI) was calculated. The high off-scale MIC was converted into the next highest concentration for calculation of the FICI, such as >16 μg/ml, was converted into 32 μg/ml. The synergy testing was repeated three times, and the higher FICI value of the two close results was reported. The interactions were defined as synergistic if the FICI was ≤0.5, indifferent if 0.5 < FICI ≤ 4.0, and antagonistic if a value of FICI was >4.0 (44).
Identification of L. prolificans Cyp51 and Fks1 orthologues through alignment.
Comparing partially published amino acid sequence of Cyp51 (GenBank accession no. MH120874.1) and Fks1 (GenBank accession no. EU337013.1) with L. prolificans strain JHH-5317 whole-genome sequence (GenBank accession no. NLAX01000008.1) plus gene annotation, we identified the Cyp51 homologous protein (GenPept accession no. PKS10573.1) in L. prolificans annotated as “similar to Eburicol 14-alpha-demethylase” and Fks1 homologous protein (GenPept accession no. PKS10859.1) annotated as “similar to 1, 3-beta-glucan synthase component Fks1” (18, 21, 45). Amino acid alignments of Cyp51 and Fks1 from published data and whole-genome sequencing analyzed data are shown in Fig. S1 in the supplemental material.
Cyp51, the promoter of Cyp51, and Fks1 amplification and sequencing.
Genomic DNA of 21 selected L. prolificans isolates from 9 patients (Table 1) (12 isolates whose MICs for azoles and micafungin were all greater than the highest MIC values and 9 isolates whose MICs for azoles and micafungin were less than or equal to the highest MIC values) was extracted from the hyphal mass according to the manufacturer’s instructions provided in Zymo Research Quick-DNA fungal/bacterial miniprep kit (Irvine, CA) and then was used as the template for amplification of the target sequences. The primers used for PCR amplification and sequencing are shown in Table S1. PrimeStar HS DNA polymerase (TaKaRa Bio, CA) was used for PCR amplification to get high-fidelity target sequences, and the PCR products were sequenced by Sanger sequencing (Thermo Fisher Scientific, Waltham, MA).
Data availability.
The L. prolificans Cyp51 sequence was deposited in GenBank with accession nos. MN329109 and MN329110. The 5′ untranscribed region (UTR) region sequence of the Cyp51 gene was deposited in GenBank with accession nos. MN329111 through MN329115. The Fks1 sequence was deposited in GenBank with accession nos. MN329116 through MN329123.
Supplementary Material
ACKNOWLEDGMENT
Yongqin Wu was supported by China Scholarship Council (no. 201806100109).
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Seidel D, Meißner A, Lackner M, Piepenbrock E, Salmanton-García J, Stecher M, Mellinghoff S, Hamprecht A, Durán Graeff L, Köhler P, Cheng MP, Denis J, Chedotal I, Chander J, Pakstis DL, Los-Arcos I, Slavin M, Montagna MT, Caggiano G, Mares M, Trauth J, Aurbach U, Vehreschild MJGT, Vehreschild JJ, Duarte RF, Herbrecht R, Wisplinghoff H, Cornely OA. 2019. Prognostic factors in 264 adults with invasive Scedosporium spp. and Lomentospora prolificans infection reported in the literature and FungiScope. Crit Rev Microbiol 45:1–21. doi: 10.1080/1040841X.2018.1514366. [DOI] [PubMed] [Google Scholar]
- 2.Guerrero A, Torres P, Duran MT, Ruiz-Diez B, Rosales M, Rodriguez-Tudela JL. 2001. Airborne outbreak of nosocomial Scedosporium prolificans infection. Lancet 357:1267–1268. doi: 10.1016/S0140-6736(00)04423-8. [DOI] [PubMed] [Google Scholar]
- 3.Rodriguez-Tudela JL, Berenguer J, Guarro J, Kantarcioglu AS, Horre R, de Hoog GS, Cuenca-Estrella M. 2009. Epidemiology and outcome of Scedosporium prolificans infection, a review of 162 cases. Med Mycol 47:359–370. doi: 10.1080/13693780802524506. [DOI] [PubMed] [Google Scholar]
- 4.Cortez KJ, Roilides E, Quiroz-Telles F, Meletiadis J, Antachopoulos C, Knudsen T, Buchanan W, Milanovich J, Sutton DA, Fothergill A, Rinaldi MG, Shea YR, Zaoutis T, Kottilil S, Walsh TJ. 2008. Infections caused by Scedosporium spp. Clin Microbiol Rev 21:157–197. doi: 10.1128/CMR.00039-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meletiadis J, Mouton JW, Meis JF, Verweij PE. 2003. In vitro drug interaction modeling of combinations of azoles with terbinafine against clinical Scedosporium prolificans isolates. Antimicrob Agents Chemother 47:106–117. doi: 10.1128/AAC.47.1.106-117.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yustes C, Guarro J. 2005. In vitro synergistic interaction between amphotericin B and micafungin against Scedosporium spp. Antimicrob Agents Chemother 49:3498–3500. doi: 10.1128/AAC.49.8.3498-3500.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meletiadis J, Mouton JW, Rodriguez-Tudela JL, Meis JF, Verweij PE. 2000. In vitro interaction of terbinafine with itraconazole against clinical isolates of Scedosporium prolificans. Antimicrob Agents Chemother 44:470–472. doi: 10.1128/aac.44.2.470-472.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li JY, Yong TY, Grove DI, Coates PT. 2008. Successful control of Scedosporium prolificans septic arthritis and probable osteomyelitis without radical surgery in a long-term renal transplant recipient. Transpl Infect Dis 10:63–65. doi: 10.1111/j.1399-3062.2007.00240.x. [DOI] [PubMed] [Google Scholar]
- 9.Gosbell IB, Toumasatos V, Yong J, Kuo RS, Ellis DH, Perrie RC. 2003. Cure of orthopaedic infection with Scedosporium prolificans, using voriconazole plus terbinafine, without the need for radical surgery. Mycoses 46:233–236. doi: 10.1046/j.1439-0507.2003.00878.x. [DOI] [PubMed] [Google Scholar]
- 10.Bhat SV, Paterson DL, Rinaldi MG, Veldkamp PJ. 2007. Scedosporium prolificans brain abscess in a patient with chronic granulomatous disease: successful combination therapy with voriconazole and terbinafine. Scand J Infect Dis 39:87–90. doi: 10.1080/00365540600786564. [DOI] [PubMed] [Google Scholar]
- 11.Rossi D, Spadari CC, Nosanchuk JD, Taborda CP, Ishida K. 2017. Miltefosine is fungicidal to Paracoccidioides spp. yeast cells but subinhibitory concentrations induce melanisation. Int J Antimicrob Agents 49:465–471. doi: 10.1016/j.ijantimicag.2016.12.020. [DOI] [PubMed] [Google Scholar]
- 12.Spadari CC, Vila T, Rozental S, Ishida K. 2018. Miltefosine has a postantifungal effect and induces apoptosis in Cryptococcus yeasts. Antimicrob Agents Chemother 62:e00312-18. doi: 10.1128/AAC.00312-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Imbert S, Palous M, Meyer I, Dannaoui E, Mazier D, Datry A, Fekkar A. 2014. In vitro combination of voriconazole and miltefosine against clinically relevant molds. Antimicrob Agents Chemother 58:6996–6998. doi: 10.1128/AAC.03212-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Compain F, Botterel F, Sitterle E, Paugam A, Bougnoux ME, Dannaoui E. 2015. In vitro activity of miltefosine in combination with voriconazole or amphotericin B against clinical isolates of Scedosporium spp. J Med Microbiol 64:309–311. doi: 10.1099/jmm.0.000019. [DOI] [PubMed] [Google Scholar]
- 15.Biswas C, Sorrell TC, Djordjevic JT, Zuo X, Jolliffe KA, Chen SC. 2013. In vitro activity of miltefosine as a single agent and in combination with voriconazole or posaconazole against uncommon filamentous fungal pathogens. J Antimicrob Chemother 68:2842–2846. doi: 10.1093/jac/dkt282. [DOI] [PubMed] [Google Scholar]
- 16.Pellon A, Ramirez-Garcia A, Buldain I, Antoran A, Martin-Souto L, Rementeria A, Hernando FL. 2018. Pathobiology of Lomentospora prolificans: could this species serve as a model of primary antifungal resistance? Int J Antimicrob Agents 51:10–15. doi: 10.1016/j.ijantimicag.2017.06.009. [DOI] [PubMed] [Google Scholar]
- 17.Sharma C, Chowdhary A. 2017. Molecular bases of antifungal resistance in filamentous fungi. Int J Antimicrob Agents 50:607–616. doi: 10.1016/j.ijantimicag.2017.06.018. [DOI] [PubMed] [Google Scholar]
- 18.Bernhardt A, Meyer W, Rickerts V, Aebischer T, Tintelnot K. 2018. Identification of 14-alpha-lanosterol demethylase (CYP51) in Scedosporium species. Antimicrob Agents Chemother 62:e02599-17. doi: 10.1128/AAC.02599-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Perlin DS. 2015. Mechanisms of echinocandin antifungal drug resistance. Ann N Y Acad Sci 1354:1–11. doi: 10.1111/nyas.12831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Katiyar SK, Edlind TD. 2009. Role for Fks1 in the intrinsic echinocandin resistance of Fusarium solani as evidenced by hybrid expression in Saccharomyces cerevisiae. Antimicrob Agents Chemother 53:1772–1778. doi: 10.1128/AAC.00020-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson ME, Katiyar SK, Edlind TD. 2011. New Fks hot spot for acquired echinocandin resistance in Saccharomyces cerevisiae and its contribution to intrinsic resistance of Scedosporium species. Antimicrob Agents Chemother 55:3774–3781. doi: 10.1128/AAC.01811-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cao D, Yao S, Zhang H, Wang S, Jin X, Lin D, Fang H, Yu Y. 2020. Mutation in cyp51A and high expression of efflux pump gene of Aspergillus fumigatus induced by propiconazole in liquid medium and soil. Environ Pollut 256:113385. doi: 10.1016/j.envpol.2019.113385. [DOI] [PubMed] [Google Scholar]
- 23.Snelders E, Camps SM, Karawajczyk A, Rijs AJ, Zoll J, Verweij PE, Melchers WJ. 2015. Genotype-phenotype complexity of the TR46/Y121F/T289A cyp51A azole resistance mechanism in Aspergillus fumigatus. Fungal Genet Biol 82:129–135. doi: 10.1016/j.fgb.2015.06.001. [DOI] [PubMed] [Google Scholar]
- 24.Mellado E, Garcia-Effron G, Alcazar-Fuoli L, Cuenca-Estrella M, Rodriguez-Tudela JL. 2004. Substitutions at methionine 220 in the 14alpha-sterol demethylase (Cyp51A) of Aspergillus fumigatus are responsible for resistance in vitro to azole antifungal drugs. Antimicrob Agents Chemother 48:2747–2750. doi: 10.1128/AAC.48.7.2747-2750.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dorlo TP, Balasegaram M, Beijnen JH, de Vries PJ. 2012. Miltefosine: a review of its pharmacology and therapeutic efficacy in the treatment of leishmaniasis. J Antimicrob Chemother 67:2576–2597. doi: 10.1093/jac/dks275. [DOI] [PubMed] [Google Scholar]
- 26.Paris C, Loiseau PM, Bories C, Breard J. 2004. Miltefosine induces apoptosis-like death in Leishmania donovani promastigotes. Antimicrob Agents Chemother 48:852–859. doi: 10.1128/aac.48.3.852-859.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pinto-Martinez AK, Rodriguez-Durán J, Serrano-Martin X, Hernandez-Rodriguez V, Benaim G. 2017. Mechanism of action of miltefosine on Leishmania donovani involves the impairment of acidocalcisome function and the activation of the sphingosine-dependent plasma membrane Ca(2+) channel. Antimicrob Agents Chemother 62:e01614-17. doi: 10.1128/AAC.01614-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Howden BP, Slavin MA, Schwarer AP, Mijch AM. 2003. Successful control of disseminated Scedosporium prolificans infection with a combination of voriconazole and terbinafine. Eur J Clin Microbiol Infect Dis 22:111–113. doi: 10.1007/s10096-002-0877-z. [DOI] [PubMed] [Google Scholar]
- 29.Raffetin A, Courbin V, Jullien V, Dannaoui E. 2017. In vitro combination of isavuconazole with echinocandins against azole-susceptible and -resistant Aspergillus spp. Antimicrob Agents Chemother 62:e01382-17. doi: 10.1128/AAC.01382-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Denardi LB, Oliveira V, de Jesus F, Dalla-Lana BH, Santurio JM, Zanette RA, Alves SH. 2018. In vitro interactions of azoles and echinocandins against clinical strains of Aspergillus flavus. Med Mycol 56:1006–1011. doi: 10.1093/mmy/myx159. [DOI] [PubMed] [Google Scholar]
- 31.Pellon A, Ramirez-Garcia A, Buldain I, Antoran A, Rementeria A, Hernando FL. 2017. Molecular and cellular responses of the pathogenic fungus Lomentospora prolificans to the antifungal drug voriconazole. PLoS One 12:e0174885. doi: 10.1371/journal.pone.0174885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sewell TR, Zhu J, Rhodes J, Hagen F, Meis JF, Fisher MC, Jombart T. 2019. Nonrandom distribution of azole resistance across the global population of Aspergillus fumigatus. mBio 10:e00392-19. doi: 10.1128/mBio.00392-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rybak JM, Fortwendel JR, Rogers PD. 2019. Emerging threat of triazole-resistant Aspergillus fumigatus. J Antimicrob Chemother 74:835–842. doi: 10.1093/jac/dky517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Albarrag AM, Anderson MJ, Howard SJ, Robson GD, Warn PA, Sanglard D, Denning DW. 2011. Interrogation of related clinical pan-azole-resistant Aspergillus fumigatus strains: G138C, Y431C, and G434C single nucleotide polymorphisms in cyp51A, upregulation of cyp51A, and integration and activation of transposon Atf1 in the cyp51A promoter. Antimicrob Agents Chemother 55:5113–5121. doi: 10.1128/AAC.00517-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dudakova A, Spiess B, Tangwattanachuleeporn M, Sasse C, Buchheidt D, Weig M, Groß U, Bader O. 2017. Molecular tools for the detection and deduction of azole antifungal drug resistance phenotypes in Aspergillus species. Clin Microbiol Rev 30:1065–1091. doi: 10.1128/CMR.00095-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Snelders E, Karawajczyk A, Schaftenaar G, Verweij PE, Melchers WJ. 2010. Azole resistance profile of amino acid changes in Aspergillus fumigatus CYP51A based on protein homology modeling. Antimicrob Agents Chemother 54:2425–2430. doi: 10.1128/AAC.01599-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Escribano P, Recio S, Pelaez T, Bouza E, Guinea J. 2011. Aspergillus fumigatus strains with mutations in the cyp51A gene do not always show phenotypic resistance to itraconazole, voriconazole, or posaconazole. Antimicrob Agents Chemother 55:2460–2462. doi: 10.1128/AAC.01358-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wei X, Chen P, Gao R, Li Y, Zhang A, Liu F, Lu L. 2017. Screening and characterization of a non-cyp51A mutation in an Aspergillus fumigatus cox10 strain conferring azole resistance. Antimicrob Agents Chemother 61:e02101-16. doi: 10.1128/AAC.02101-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pérez-Cantero A, López-Fernández L, Guarro J, Capilla J. 2019. New insights into the Cyp51 contribution to azole resistance in Aspergillus section Nigri. Antimicrob Agents Chemother 63:e00543-19. doi: 10.1128/AAC.00543-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gonzalez-Lara MF, Roman-Montes CM, Diaz-Lomeli P, Rangel-Cordero A, Valenzuela MO, Ponce-de-Leon A, Sifuentes-Osornio J, Ostrosky-Zeichner L, Martinez-Gamboa A. 2019. Azole resistance and cyp51A mutation screening in Aspergillus fumigatus in Mexico. J Antimicrob Chemother 74:2047–2050. doi: 10.1093/jac/dkz121. [DOI] [PubMed] [Google Scholar]
- 41.Ballard E, Weber J, Melchers W, Tammireddy S, Whitfield PD, Brakhage AA, Brown A, Verweij PE, Warris A. 2019. Recreation of in-host acquired single nucleotide polymorphisms by CRISPR-Cas9 reveals an uncharacterised gene playing a role in Aspergillus fumigatus azole resistance via a non-cyp51A mediated resistance mechanism. Fungal Genet Biol 130:98–106. doi: 10.1016/j.fgb.2019.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aruanno M, Bachmann D, Sanglard D, Lamoth F. 2019. Link between heat shock protein 90 and the mitochondrial respiratory chain in the caspofungin stress response of Aspergillus fumigatus. Antimicrob Agents Chemother 63:e00208-19. doi: 10.1128/AAC.00208-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Satish S, Jimenez-Ortigosa C, Zhao Y, Lee MH, Dolgov E, Kruger T, Park S, Denning DW, Kniemeyer O, Brakhage AA, Perlin DS. 2019. Stress-induced changes in the lipid microenvironment of beta-(1,3)-d-glucan synthase cause clinically important echinocandin resistance in Aspergillus fumigatus. mBio 10:e00779-19. doi: 10.1128/mBio.00779-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Odds FC. 2003. Synergy, antagonism, and what the chequerboard puts between them. J Antimicrob Chemother 52:1. doi: 10.1093/jac/dkg301. [DOI] [PubMed] [Google Scholar]
- 45.Luo R, Zimin A, Workman R, Fan Y, Pertea G, Grossman N, Wear MP, Jia B, Miller H, Casadevall A, Timp W, Zhang SX, Salzberg SL. 2017. First draft genome sequence of the pathogenic fungus Lomentospora prolificans (formerly Scedosporium prolificans). G3 (Bethesda) 7:3831–3836. doi: 10.1534/g3.117.300107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The L. prolificans Cyp51 sequence was deposited in GenBank with accession nos. MN329109 and MN329110. The 5′ untranscribed region (UTR) region sequence of the Cyp51 gene was deposited in GenBank with accession nos. MN329111 through MN329115. The Fks1 sequence was deposited in GenBank with accession nos. MN329116 through MN329123.