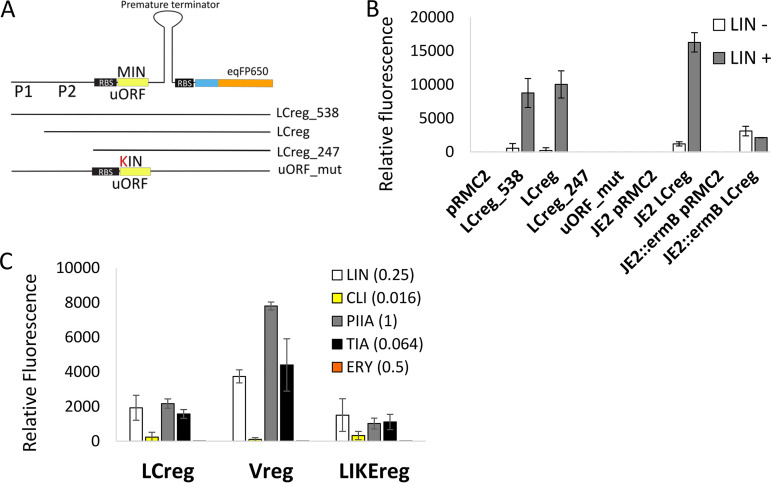
FIG 2.
Expression of vga(A) is regulated by a ribosome-mediated attenuation mechanism. (A) Testing the activity of transcriptional attenuator encoded upstream of vga(A)LC using fluorescent reporter fusion. Graphical overview of reporter constructs encompassing 538, 375, and 247 nucleotides upstream of the vga(A)LC, including predicted promoters P1 and P2 and the first 19 codons fused to eqFP650 fluorescent protein and LCreg_538 construct in which the start codon ATG of the predicted upstream regulatory open reading frame (uORF), was mutated to AAG (uORF_mut). (B) Relative fluorescence intensity of eqFP650 reporter constructs LCreg_538, LCreg, LCreg_247, and uORF_mut expressed in S. aureus RN4220 in the absence or the presence of lincomycin (LIN, 0.25 mg/liter) and LCreg reporter construct expressed in S. aureus JE2 without or with constitutively expressed ermB, inserted in the genome. ErmB prevents the binding of lincomycin to the ribosome. (C) Relative fluorescence intensity of S. aureus RN4220 expressing eqFP650 fluorescent protein under the control of vga(A)LC (LCreg), vga(A)V (Vreg), or vga(A)LIKE (LIKEreg) 5′ UTRs in response to lincomycin (LIN), clindamycin (CLI), pristinamycin IIA (PIIA), tiamulin (TIA), and erythromycin (ERY) at an antibiotic concentration which corresponded to the maximum level of induction. (B, C) Averages and standard deviations of three independent measurements are shown. The alignment of 5′ UTRs and secondary structure predictions are shown in Fig. S5 in the supplemental material. The full range of antibiotic concentrations and corresponding fluorescence levels are shown in Fig. S6a. The growth of S. aureus RN4220 in the presence of antibiotics is shown in Fig. S6b.