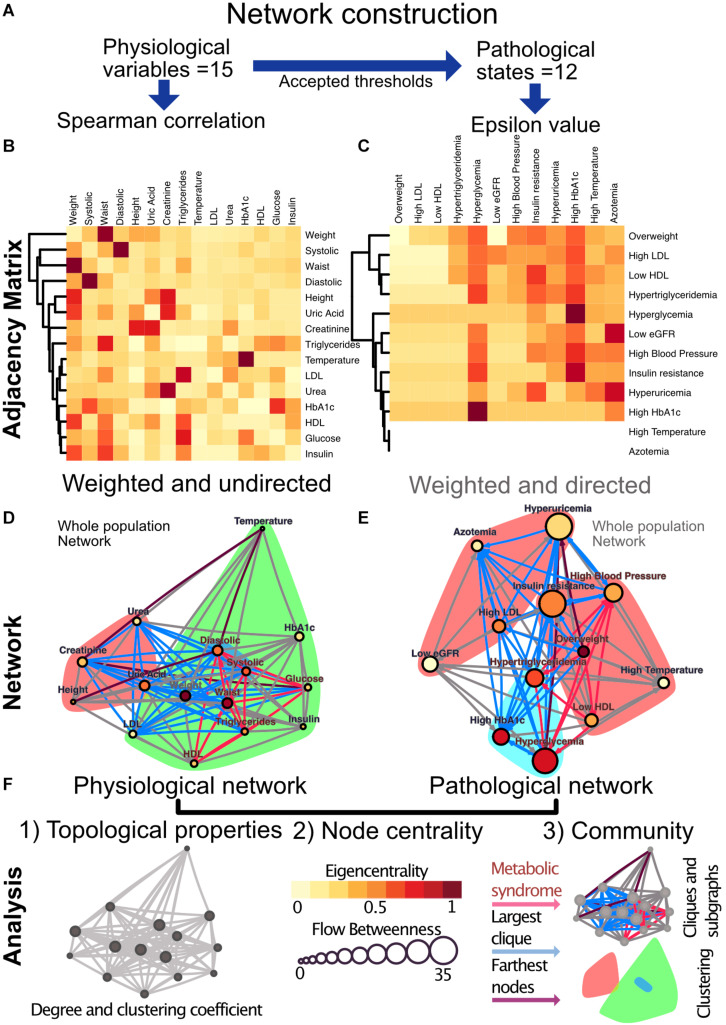
FIGURE 4.
Metabolic physiological network construction from matrices. (A) Correlation of 15 physiological variables and their corresponding 12 pathological states associations were modeled using Spearman correlation and ε value, respectively. (B) Adjacency matrix as a heatmap where the darker the red indicates a greater monotonic relationship between two physiological variables, as calculated by the Spearman rank correlation rho. (C) ε Value between each pair of pathological states, a darker red indicating a greater probability of coexistence. In both heatmaps, rows and columns are ordered by weighted degree, and on the left side of the heat maps the resulting hierarchical dendrogram is shown. For directed networks some nodes lacked outgoing links, this is presented as blank rows. (D) Undirected network of physiological variables for the whole sample. The edges are weighted by the rho value in the Spearman correlation. The size of the node shows the flow betweenness of a node, the eigencentrality is shown by its color and the color shadowed areas indicate the Louvain clusters. (E) Directed network of pathological states. The edges are weighted by the ε value, the size of the node shows the flow betweenness of each node, the eigencentrality is shown by its color and the color shadowed areas indicate spinglass clusters. (F) In both networks, the red subgraph shows the components of MetS, while the blue subgraph highlights the largest clique and the diameter of the network is in purple. For Spearman correlation, values with p > 0.001 were discarded, whereas for ε, values below 1.96 were discarded.