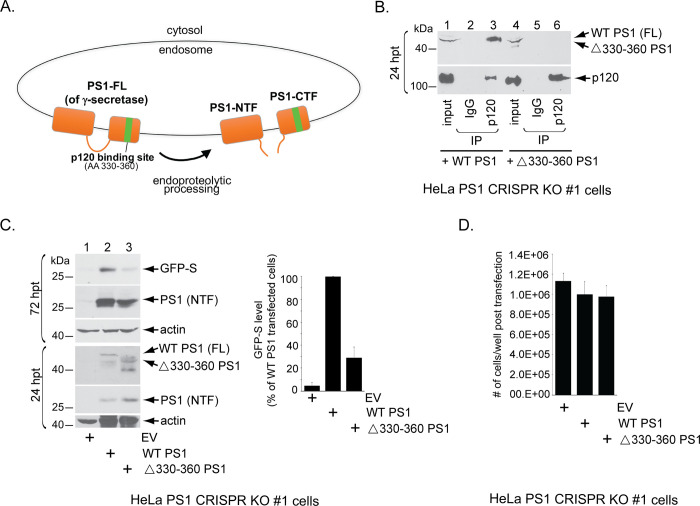
Fig 2. A γ-secretase mutant that cannot bind to p120 inefficiently supports HPV16 infection.
A. A model depicting the p120-binding site in the PS1 subunit of γ-secretase. p120 binds to amino acid (AA) 330–360 of PS1. “FL”, full-length; “NTF”, N-terminal fragment; “CTF”, C-terminal fragment B. HeLa PS1 CRISPR knockout cells were transfected with a plasmid expressing either WT PS1 or PS1 deleted of amino acids 330–360 (Δ330–360 PS1). 24 hrs post-transfection (hpt), cells were lysed and the resulting extract was subjected to immunoprecipitation using either a control IgG antibody or an antibody against p120. The precipitated material was subjected to SDS-PAGE and immunoblotting using antibodies recognizing p120 or FLAG-tagged L2. Only the full-length (FL) PS1 (prior to endoproteolytic processing) is shown. C. (Upper panels, 72 hrs post-transfection, hpt) HeLa PS1 CRISPR knockout cells were transfected with empty vector (EV), WT PS1, or Δ330–360 PS1 for 24 hrs, and then infected with WT HPV16.L2F (GFP-S). 48 hpi, cells were lysed and the resulting extract subjected to SDS-PAGE and immunoblotting using the indicated antibodies. The levels of GFP-S were quantified as in Fig 1. Data are normalized against infected cells transfected with WT PS1, and further normalized against the actin levels. Data represent the mean ± SD of three independent experiments. (Lower panels, 24 hrs post-transfection, hpt) A pool of cells were harvested 24 hrs post-transfection with the indicated DNA in the absence of PsV. The resulting cell extract was subjected to SDS-PAGE and immunoblotting using antibodies recognizing the indicated proteins to visualize protein levels at the time of infection. Both the full-length PS1 and the N-terminal fragment of PS1 (due to endoproteolytic processing) are observed at the 24 hpt time point. D. HeLa PS1 CRISPR KO #1 cells were seeded at equal amounts in a 6-well plate and transfected with empty vector (EV), WT PS1, or Δ330–360 PS1 for 24 hrs. 72 hours after transfection, cells were harvested and the total number of cells per condition were counted by hemocytometer. Data represent the mean ± SD of three independent experiments.