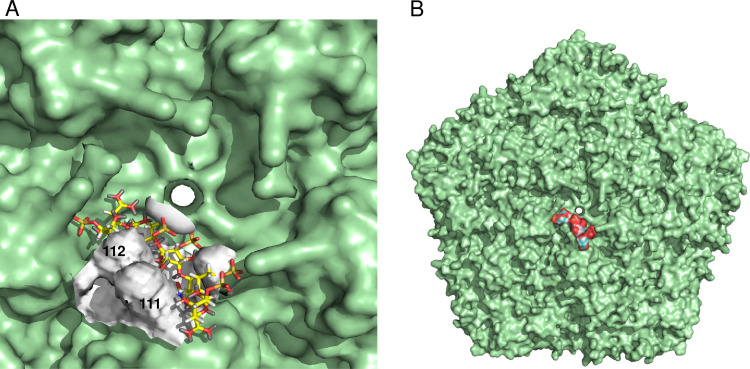
Fig 2. GRID [41] was used to find the energetically favorable binding site for HSPG on the SAT1 modelled mutant capsid.
(A) The GRID calculation was performed for a 20 Å radius around the 5-fold axis using pyramidal sulfur as a probe. VP1 residue 112 is the most likely site of interaction with molecular interaction energy of -8.2 kcal/mole. The interaction energy increased to -10 kcal/mole when the grid was centered at VP1 residue 112. (B) Five linked heparin disaccharide molecules were docked using the default parameters of GOLD onto the SAT1 modeled mutant pentamer structures. A 30Å3 region from VP1 residue 112 was defined for docking and the GOLD fitness score function was used to rank the docking poses. The best docking pose is shown (GOLD score = 127). The equivalent process for the wild-type virus produced a less satisfactory docking (GOLD score = 102, docking not shown).