Abstract
Objective
To test the hypothesis that genetic risk for Alzheimer disease (AD) may represent a stable influence on the brain from early in life, rather than being primarily age dependent, we investigated in a lifespan sample of 1,181 persons with a total of 2,690 brain scans, whether higher polygenic risk score (PGS) for AD and presence of APOE ε4 was associated with lower hippocampal volumes to begin with, as an offset effect, or possibly faster decline in older age.
Methods
Using general additive mixed models, we assessed the relations of PGS for AD, including variants in APOE with hippocampal volume and its change in a cognitively healthy longitudinal lifespan sample (age range: 4–95 years, mean visit age 39.7 years, SD 26.9 years), followed for up to 11 years.
Results
AD-PGS and APOE ε4 in isolation showed a significant negative effect on hippocampal volume. The effect of a 1 sample SD increase in AD-PGS on hippocampal volume was estimated to –36.4 mm3 (confidence interval [CI]: –71.8, –1.04) and the effect of carrying ε4 allele(s) –107.0 mm3 (CI: –182.0, –31.5). Offset effects of AD-PGS and APOE ε4 were present in hippocampal development, and interactions between age and genetic risk on volume change were not consistently observed.
Conclusions
Endophenotypic manifestation of polygenic risk for AD may be seen across the lifespan in cognitively healthy persons, not being confined to clinical populations or older age. This emphasizes that a broader population and age range may be relevant targets for attempts to prevent AD.
Among the earliest signs of Alzheimer disease (AD) are deficits in memory and orientation, tied to hippocampal dysfunction.1 Although variants at multiple genetic loci identified in case-control studies are associated with AD risk,2 little is known about how these affect individuals devoid of AD diagnosis at an endophenotypic level. Although the major AD genetic risk factor, the APOE ε4 allele,3,4 has been associated with smaller hippocampi at various ages also in healthy persons,5,6 longitudinal data are scarce and restricted to older adults.7,8 It is unknown whether polygenic risk scores (PGSs) calculated from established AD risk variants translate to differences in neural characteristics at different life stages in healthy persons. Of interest, higher AD-PGS was recently found related to lower left hippocampal volumes in young adults, also when variants in APOE were excluded.6 The negative effect of common genetic polymorphisms on late-life disease, including AD, has often been interpreted in terms of age-specific mechanisms.9,10 However, there is growing evidence for continuous influence of early life factors on later-life cognitive function and neural foundations.11–14 Do AD genetic risk factors exert their effects only at later life stages resulting in brain atrophy and clinical symptoms or affect neural structures throughout the lifespan in the population at large? We hypothesized a relation throughout the lifespan, with higher AD-PGS, including the presence of APOE ε4, showing association with lower hippocampal volumes early in life, as an offset effect. We tested whether AD genetic risk factors derived from genome-wide association studies2 had an effect on hippocampal volume and volumetric changes in cognitively healthy individuals through the lifespan and whether genetic risk interacted with age.
Methods
Sample
A total of 2,690 valid scans from 1,181 cognitively healthy participants, aged 4.1–95.7 years (mean visit age 39.7 years, SD 26.9 years), were drawn from 5 Norwegian studies. The studies included 4 substudies coordinated by the Center for Lifespan Changes in Brain and Cognition (LCBC); The Norwegian Mother and Child Cohort Neurocognitive Study,15 Neurocognitive Development,16 Cognition and Plasticity Through the Lifespan,17 Neurocognitive Plasticity,18 and a study run collaboratively by LCBC and Oslo University Hospital, Novel Biomarkers19 (see e-Methods for details, links.lww.com/NXG/A316). The majority of participants were followed longitudinally, scan intervals ranging 0.2–11.0 years (mean = 2.9 years, SD = 2.6 years). The sample is partly overlapping with a lifespan sample used in previous reports.12,14 All were screened, and dementia, previous stroke with sequela, Parkinson disease, and other neurodegenerative diseases likely to affect cognition were the exclusion criteria, with additional inclusion and exclusion criteria being applied per study (see e-Methods). Sample descriptions for the total sample binned by time points are given in table 1. Additional descriptions including distribution of substudy samples per time point are given in table e-1.
Table 1.
Sample descriptives
To check for possible recruitment bias/age selectivity, the correlations of age and AD-PGS and APOE ε4 allelic variation (coded as both 0, 1, or 2 copies of the ε4 allele and the absence or presence of the ε4 allele, 0 or 1) were calculated. For AD-PGS and age, there was a modest correlation, r = −0.04 (confidence interval [CI]: −0.08, −0.00), suggesting some age selectivity. No correlations reaching statistical significance or trends toward such were observed for APOE ε4 allelic variation, correlations with age being r = −0.01 (CI: −0.05, 0.02) for APOE ε4 coded as no (0)/yes (1) and r = 0.00 (CI: −0.04, 0.04) for APOE coded as no ε4 allele (0)/1 ε4 allele (1)/2 ε4 alleles (2). This suggests that there was no substantial sample selection age bias with regard to genetic risk for AD. The proportion of APOE ε4 carriers in the present study appears largely in line with that which would be expected for the population in a Scandinavian country.20
Standard protocol approvals, registrations, and patient consents
The studies were approved by the Regional Ethical Committee of South East Norway. Written informed consent was obtained from participants aged 16 years and older and from parents of participants younger than 16 years, and oral consent was obtained from participants aged 12 years and older.
Genotyping
Buccal swab and saliva samples were collected for DNA extraction, followed by genome-wide genotyping using the Global Screening Array (Illumina, Inc., San Diego, CA). For a full description of genotyping and postgenotyping methods, including quality control and imputation of untyped markers, please see e-Methods (links.lww.com/NXG/A316). AD-PGS of our sample was calculated using the allelic effect sizes from Lambert et al.2 Shared single nucleotide polymorphisms (SNPs) between our data and Lambert et al.2 were pruned to be nearly independent using the program PLINK21 with the following parameters --clump-p1 1.0 --clump-p2 1.0 --clump-kb 500 --clump-r2 0.1. The linkage disequilibrium structure was based on the European subpopulation from the 1000 Genomes Project Phase 3.22 Because of the complexity of the major histocompatibility complex region (build hg19; chr6:25652429-33,368,333), we removed SNPs in this region except the most significant one before pruning. Previous studies23–25 have shown that PGSs constructed using SNPs with association p value <0.5 from Lambert et al.2 have the largest effect on the risk of AD. Hence, we used the same threshold in the pruned set for computing the AD-PGS. To investigate the consistency of results across different p value thresholds, SNPs with p < 5e-08 in the pruned set were additionally used for constructing a PGS for our samples. Analyses were recomputed limiting markers to those showing genome-wide significant association (i.e., p < 5e-08) with AD risk in Lambert et al.2 Because of its known large effect, we computed AD-PGS with and without markers in the APOE region (build hg19; chr19:44,909,011-45,912,650). To test the effect of APOE itself, we modeled the counts of APOE ε4 alleles directly by counting the number of alternative alleles at rs429358,26,27 as 0, 1, or 2. To control the possible effect of vascular factors on the relation between AD-PGS and hippocampus volume, we included PGSs for systolic and diastolic blood pressure (SBP-PGS and DBP-PGS) as additional covariates in our models. Both SBP-PGS and DBP-PGS were computed using the same protocol as that used for AD-PGS, but based on the public data from Liu et al.28 We computed the genetic ancestry factors (GAFs) using principal component methods.29 For the present analysis, only participants of European ancestry were included, excluding 89 persons for whom we had genotype data.
MRI data acquisition
Participants were scanned at a total of 4 Siemens scanners at 2 sites. The pulse sequences used for morphometric analyses were 1 to 2 3D sagittal T1-weighted MPRAGE sequences. For details on the scanners and pulse sequences used, see e-Methods (links.lww.com/NXG/A316). Other MRI volumes were recorded, including sequences intended for and examined by a radiologist, to rule out and medically follow up incidental neuroradiologic findings. Distribution of scans from the different scanners per time point is given in table e-1. See e-Methods for further details.
Image analysis
All scans were reviewed for quality and automatically corrected for spatial distortion.30 Images were processed with the FreeSurfer software package (version 6.0), including automatic hippocampal volumetric segmentation.31 To allow assessment of differences between scanners, 24 participants were scanned on all 3 scanners from Oslo University Hospital on the same day. Linear regression analyses were run testing the concordance between hippocampal volumes between scanners, yielding excellent agreement. Thus, including scanner as a covariate in the analyses would account for offset differences between scanners. See e-Methods for further details (links.lww.com/NXG/A316).
Statistical analyses
Analyses were run in R version 3.6.0. General additive mixed models using the package mgcv version 1.8-28 were used to derive age functions with a random intercept term per participant. Hippocampal volumes were predicted from (1) a smooth function of age and a linear function of AD-PGS, (2) a smooth function of age and linear functions of AD-PGS and presence/absence of 1 or 2 APOE ε4 alleles, (3) smooth functions of age and AD-PGS and a tensor interaction term of age and AD-PGS, and (4) a linear function of the presence/absence of 1 or 2 APOE ε4 alleles and a smooth interaction between age and presence/absence of 1 or 2 APOE ε4 alleles. In all models, scanner, intracranial volume, sex, and the first 5 GAFs, in addition to genotyping batch (1; n = 1,014, 2; n = 166), were entered as covariates. DBP-PGS and SBP-PGS were added as additional covariates (repeating steps 2 and 4), to further investigate whether any effect was unique to AD-PGS and APOE. We did not have any hypothesis regarding differential hemispheric effects, and bilateral hippocampal volumes were entered in analyses. However, we additionally ran analyses for the left and right hippocampi separately (repeating steps 2 and 4), and to allow for comparison with others,6 results presented as e-Methods (links.lww.com/NXG/A316). We computed 95% CIs for the effect of PGS and APOE allelic variation. The reported effect sizes are mm3 difference in hippocampal volume per SD of PGS and when carrying 1 or more ε4 alleles vs not carrying any.
Data availability
There are data access restrictions in accordance with ethical guidelines and national data protection legislation. The data will be made available pending reasonable request, appropriate institutional data protection security measures, and ethical approval.
Results
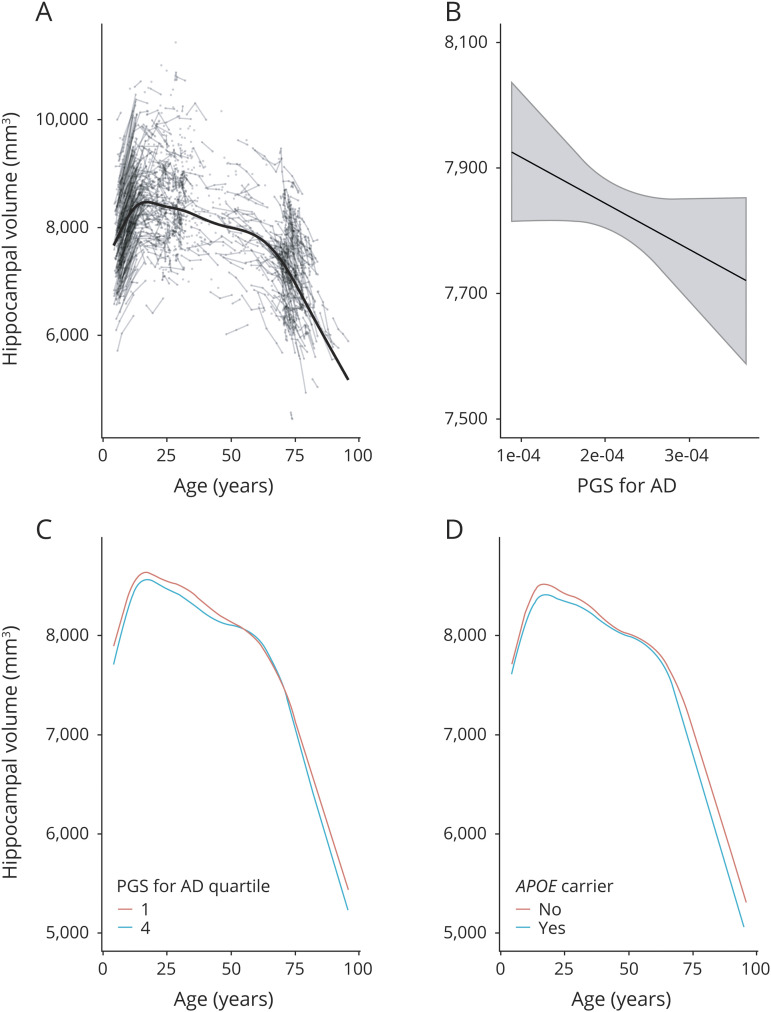
In the lifespan sample, hippocampal volumes increased in early development and declined in older age, as shown in figure 1A. We found a significant negative effect of AD-PGS on hippocampal volume, as shown in figure 1B. The estimated effect of AD-PGS on hippocampal volume was −36.4 mm3 (CI: −71.8, −1.0) associated with 1 sample SD higher AD-PGS. The interaction term between age and AD-PGS did not reach statistical significance. The interaction term between age and AD-PGS represents how the effect of AD-PGS varies with age and is nonlinear in both variables. The point estimate of the interaction was less than 10% of the main effect of AD-PGS for AD-PGS within 2 SDs of the sample mean over the full age range considered. At age 20 years, this interaction term estimated an additional offset effect −0.27 mm3 (CI: −2.87, 2.33) at AD-PGS 1 SD below the mean and 0.23 mm3 (CI: −2.39, 2.85) at 1 SD above the mean, and at age 80 years, the corresponding estimates were 0.96 mm3 (CI: −3.12, 5.04) and −0.86 mm3 (CI: −4.96, 3.24). Of note, this implies that the absolute volume difference between those with lower and higher AD-PGS appears to be about the same in young adults (∼20s) as in the oldest adults (∼80s), and even the extreme ends of the CIs represent quite moderate differences. Although the analysis was based on the continuous AD-PGS score, inspection of the age trajectories for values of AD-PGS at 1 SD below and above mean (figure 1C) did not give any consistent indication of greater effects of higher PGS for AD with age. Rather, although there was time after age 60 years when the hippocampal curves merge for the different subgroups, they were otherwise mostly ordered throughout life so that those with PGS for AD 1 SD below mean had the highest volume, whereas those with the PGS for AD 1 SD below mean have the lowest volume. Adding PGSs for blood pressure (DBP and SBP) as additional covariates, the effect of AD-PGS on hippocampal volume was still significant, see e-Methods for further details (links.lww.com/NXG/A316).
Figure 1. Hippocampal volume in relation to age and genetic risk.
Hippocampal volume (across hemispheres, shown in mm3 on the y-axis) and change in relation to (A) age (in years, x-axis) plotted with individual trajectories overlaid, (B) polygenic risk score (PGS) for Alzheimer disease (AD) (x-axis, continuous scale 0–1), (C) age (in years, x-axis) with PGS for AD set to the first (red line) and fourth quartiles (blue line) of the sample, and (D) trajectories for carriers (blue line) and noncarriers (red line) of the APOE ε4 allele. PGSs for AD shown here were constructed using single nucleotide polymorphisms with association p value <0.5 from Lambert et al.2
Analysis with APOE status as the predictor of hippocampal volume likewise showed a significant negative effect of the presence of the ε4 allele. The estimated effect of carrying 1 or more ε4 alleles on hippocampal volume was −107.0 mm3 (CI: −182.0, −31.5). The interaction term between age and APOE allelic variance did not reach statistical significance. ε4 allele carriers had smaller hippocampal volumes in young age, as an offset effect. There was not indication of faster atrophy in older age, as carriers in young adulthood (∼20s) appeared to display lower hippocampal volumes, a difference that seemed somewhat lower in middle age, but more pronounced again in older adulthood (∼80s; figure 1D). At age 20 years, the difference in hippocampal volume between carriers and noncarriers was −101 mm3 (CI: −224, 23); at age 50 years, this difference was −22.6 mm3 (CI: −186, 141); and at age 80 years, it was −278 mm3 (CI: −478, −77.7). Adding PGSs for blood pressure (DBP and SBP) as additional covariates did not weaken the effect of APOE status on hippocampal volume, see e-Methods for further details (links.lww.com/NXG/A316).
AD-PGS excluding the APOE region did not show a significant effect on hippocampal volume, although a trend in the same direction as the full model was observed (estimate −31.8 mm3, CI: −67.4, 3.92). There was no evidence for an age interaction on using the AD-PGS excluding markers in the APOE region. The estimated interactions corresponding to values of AD-PGS excluding the APOE region at 1 SD below the sample mean varied from −0.1 mm3 (CI: −1.1, 2.2) at age 20 years to 0.2 mm3 (CI: −2.2, 2.6) at age 80 years, and the corresponding values at other ages were very similar. For values of AD-PGS excluding the APOE region at 1 SD above the sample mean, the interaction varied from 0.1 mm3 (CI: −1.4, 1.6) at age 20 years to −0.1 mm3 (CI: −2.6, 2.3) at age 80 years, and also here the corresponding values at other ages were very similar.
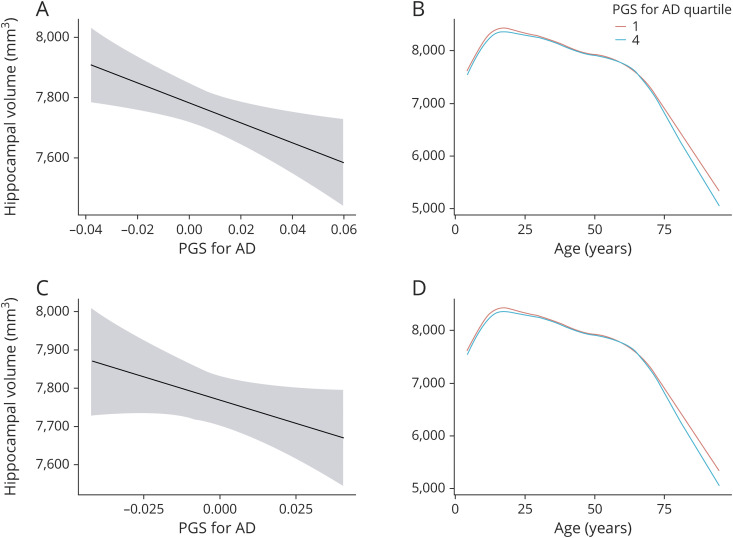
Recomputing the analyses with an AD-PGS limited to SNPs only showing genome-wide significant association (i.e., p < 5e-08) with AD risk in Lambert et al.2 confirmed a significant negative effect of AD-PGS on hippocampal volume (−44.8 mm3/AD-PGS SD, CI: −80.7, −8.9). This result was reduced to a trend when excluding markers in the APOE region (−35.6 mm3/PGS SD, CI: −76.7, 5.5). However, significant age interactions appeared in the PGS analyses limited to genome-wide significant markers, both with and without markers in the APOE region (figure 2). Especially when excluding the APOE region, there seemed to be a somewhat more negative effect of higher AD-PGS on hippocampal volume in older age (above 80 years). For a more detailed description of these interaction effects, see e-Methods (links.lww.com/NXG/A316).
Figure 2. Hippocampal volume and PGS for AD limited to SNPs showing genome-wide significant association.
Upper panel: including APOE. Hippocampal volume (across hemispheres, shown in mm3 on the y-axis) and change in relation to (A) AD-PGS including the APOE region as calculated based on effect sizes from Lambert et al.2 at p < 5e-08 (x-axis, continuous scale 0–1). (B) Age (in years, x-axis) with AD-PGS at p < 5e-08 including the APOE region set to the first (red line) and fourth (blue line) quartiles of the sample. Lower panel: excluding APOE. Hippocampal volume (across hemispheres, shown in mm3 on the y-axis) and change in relation to (C) AD-PGS excluding the APOE region as calculated based on effect sizes from Lambert et al.2 at p < 5e-08 (x-axis, continuous scale 0–1). (D) Age (in years, x-axis) with AD-PGS at p < 5e-08 excluding the APOE region set to the first (red line) and fourth (blue line) quartiles of the sample. AD = Alzheimer disease; PGS = polygenic risk score; SNP = single nucleotide polymorphism.
Discussion
This study shows that genetic risk for AD is associated with effects on hippocampal volume throughout life, having neurodevelopmental offset effects observable from childhood in cognitively healthy well-functioning participants. Higher AD-PGS and carrying the APOE ε4 allele specifically were associated with lower developmental hippocampal volume offsets, and persons with higher genetic risk for AD remained having lower hippocampal volumes with age. Notably, this main effect was consistently observed for PGSs constructed using SNPs with 2 different association p values.2 The association of hippocampal volume and AD-PGS through the lifespan found here can contribute to build knowledge on the lifespan trajectories of polygenic AD risk at an endophenotypic level. However, and as expected, the effect was not large, and when using the AD-PGSs computed without the APOE region as predictor, the effect was reduced to a trend.
A negative cross-sectional association between hippocampal volumes and polygenic risk for AD—also excluding the APOE region—has been previously reported in a smaller sample of young adults,6 suggesting that the relationship between hippocampal volume and AD risk results from multiple genetic factors and not exclusively variability in the APOE region. We do not consider those findings in conflict with the present, where a trend toward the same was observed. However, as can be seen from the associations reported, the effect of APOE on hippocampal volumes in cognitively healthy persons of varying ages appears stronger than that of other genetic variants.
Having low vs high genetic risk for AD was associated with roughly equivalent difference in hippocampal volume at age 25 and 80 years. In the current study, the effect of the PGS constructed using SNPs with association p value <0.5 from Lambert et al.,2 shown to have the largest effect on the risk of AD,23–25 did not significantly interact with age. However, the PGS effect was not equally apparent at all ages among older adults, and notably, age interactions were observed for the same PGS constructed using SNPs with the lowest association p value, also when excluding variants in the APOE region. Possibly, different extent of effects at different ages may illuminate why one small recent study found associations of an AD genetic risk score and hippocampal atrophy over a 2-year interval in healthy older adults (n = 45), although no association with individual variation in volume was seen at baseline (n = 66).8 AD-PGS did correlate modestly with age in our sample, and there may be some sample selection bias in healthy older age samples, so it would be interesting to see whether age interactions could be consistently observed if persons were followed for even longer and at higher ages. It should be noted that a limitation of the current study is that sample density was lower in the middle of the lifespan than in young and older age groups. We cannot, based on the current sample, exclude a mixture of neurodevelopmental offset and aging effects, although the neurodevelopmental offset effects were the most consistently supported by the present results.
There was only a trend for effects of APOE allelic variants to interact with age, and this trend was not clearly indicative of faster atrophy for APOE ε4 carriers in older age. Cross-sectional studies have shown that APOE ε43,4 carriers tend to have lower hippocampal/medial temporal lobe volumes at various ages also in healthy persons,5 and notably, this has been identified even in neonates.32 However, it has been difficult to interpret whether these developmental structural brain differences actually do represent long-term risk factors, as longitudinal imaging data have been lacking. The only such study we know of was restricted to older adults aged 55–75 years,7 showing greater hippocampal atrophy in ε4 carriers across a 5-year interval. The present study hence partly confirms, yet nuances and extends previous reports, in showing a main effect through the lifespan, i.e., also an offset effect.
The present study adds to the evidence for early life factors exerting a continuous influence on later-life function,11 showing that genetic factors established to influence late-life neural and cognitive disease work in part in a temporally stable and dimensional manner. That is, genetic risk factors for AD seem not to only manifest at late life in hippocampal atrophy and clinical symptoms but appear to start influencing the neural structures on which cognition rely early on, through the entire lifespan and in the population at large. In this regard, currently observed effects of the APOE ε4 allele cannot well be interpreted only within the framework of antagonistic pleiotropy,9,10 as this allelic variant appears to have in part similar effects on neural structures of memory function at different ages. As for AD-PGS, currently observed effects are of similar magnitude in young adulthood as in much of older adulthood, meaning that explanations evoking different effects on the hippocampus at different ages may be incomplete.
This does not necessarily imply that effects on behavior may be readily observed through the lifespan. A number of studies have indeed not observed effects of APOE status on standard neuropsychological memory tests.33,34 However, as recently reported, this does not mean that effects of genetic risk for AD do not manifest early, in more fine-grained behaviors dependent on hippocampal circuitry, such as spatial navigation.34 A related yet different account is the magnification or resource modulation hypothesis,35–37 stating that genetic effects are magnified in persons with constrained neural resources, such as older—and putatively also developing—individuals. Here, it is assumed that the function relating brain resources to cognition is nonlinear so that genetic differences exert increasingly larger effects on performance with lesser neural substrates. Smaller hippocampi can be seen as a risk factor. The idea that persons starting with larger brain structures may preserve mental status despite developing AD plaques, and may be said to have a greater reserve, is supported by older postmortem and clinical data.38 Within a brain reserve account, such differences in brain structure may relate to differences in tolerance to pathology before one falls under a functional threshold.39 Hence, these differences may become readily functionally apparent only in older age, but they likely are there to begin with and throughout life in a stable manner. This supports a lifespan model of dementing disorder,40 where effects of common genetic variants in part work in a stable manner to be one of several factors affecting risk for cognitive decline and neurodegenerative disease.
In conclusion, endophenotypic expression of genetic risk for AD may be seen in a dimensional and lifespan perspective, not being confined to clinical populations or older age. This emphasizes that a broader population and age range may be relevant targets for attempts to prevent AD. To help define targets for prevention, future studies of the genetic factors established to influence AD are needed. These should investigate which of these factors have an age-independent influence, across manifestations of health and disease, and which may have more pronounced effects in aging.
Acknowledgment
The authors are grateful to all participants for their time and commitment and to all colleagues in LCBC for taking part in the gathering and preprocessing of the data. They thank Mrs. Tanja Wesse, Mrs. Sanaz Sedghpour Sabet, and Dr. Michael Wittig at the Institute of Clinical Molecular Biology, Christian-Albrechts-University of Kiel, Germany, for technical assistance with the GSA genotyping. The LIGA team acknowledges computational support from the OMICS compute cluster at the University of Lübeck.
Glossary
- AD
Alzheimer disease
- CI
confidence interval
- DBP
diastolic blood pressure
- GAF
genetic ancestry factor
- LCBC
Center for Lifespan Changes in Brain and Cognition
- PGS
polygenic risk score
- SBP
systolic blood pressure
- SNP
single nucleotide polymorphism
Appendix. Authors
Study funding
This research was funded by grants from the Norwegian Research Council (to K.B.W., A.M.F., and Y.W.), the National Association for Public Health's dementia research program, Norway (to A.M.F.), and the European Research Council's Starting (grant agreement 283634 to A.M.F. and 313440 to K.B.W.) and consolidator Grant Scheme (grant agreement 771355 to K.B.W. and 725025 to A.M.F.). The funding sources had no role in the study design; in the collection, analysis, and interpretation of the data; in the writing of the report; or in the decision to submit the article for publication.
Disclosure
The authors report no disclosures. Go to Neurology.org/NG for full disclosures.
References
- 1.Chincarini A, Sensi F, Rei L, et al. . Integrating longitudinal information in hippocampal volume measurements for the early detection of Alzheimer's disease. NeuroImage 2016;125:834–847. [DOI] [PubMed] [Google Scholar]
- 2.Lambert JC, Ibrahim-Verbaas CA, Harold D, et al. . Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer's disease. Nat Genet 2013;45:1452–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Corder EH, Saunders AM, Strittmatter WJ, et al. . Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer's disease in late onset families. Science 1993;261:921–923. [DOI] [PubMed] [Google Scholar]
- 4.Genin E, Hannequin D, Wallon D, et al. . APOE and Alzheimer disease: a major gene with semi-dominant inheritance. Mol Psychiatry 2011;16:903–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Piers RJ. Structural brain volume differences between cognitively intact ApoE4 carriers and non-carriers across the lifespan. Neural Regen Res 2018;13:1309–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Foley SF, Tansey KE, Caseras X, et al. . Multimodal brain imaging reveals structural differences in Alzheimer's disease polygenic risk carriers: a study in healthy young adults. Biol Psychiatry 2017;81:154–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lu PH, Thompson PM, Leow A, et al. . Apolipoprotein E genotype is associated with temporal and hippocampal atrophy rates in healthy elderly adults: a tensor-based morphometry study. J Alzheimers Dis 2011;23:433–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harrison TM, Mahmood Z, Lau EP, et al. . An Alzheimer's disease genetic risk score predicts longitudinal thinning of hippocampal complex subregions in healthy older adults. eNeuro 2016;3:ENEURO.0098-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Han SD, Bondi MW. Revision of the apolipoprotein E compensatory mechanism recruitment hypothesis. Alzheimers Dement 2008;4:251–254. [DOI] [PubMed] [Google Scholar]
- 10.Zetterberg H, Alexander DM, Spandidos DA, Blennow K. Additional evidence for antagonistic pleiotropic effects of APOE. Alzheimer Dement 2009;5:75. [DOI] [PubMed] [Google Scholar]
- 11.Kovacs GG, Adle-Biassette H, Milenkovic I, Cipriani S, van Scheppingen J, Aronica E. Linking pathways in the developing and aging brain with neurodegeneration. Neuroscience 2014;269:152–172. [DOI] [PubMed] [Google Scholar]
- 12.Walhovd KB, Krogsrud SK, Amlien IK, et al. . Neurodevelopmental origins of lifespan changes in brain and cognition. Proc Natl Acad Sci USA 2016;113:9357–9362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Walhovd KB, Fjell AM, Brown TT, et al. . Long-term influence of normal variation in neonatal characteristics on human brain development. Proc Natl Acad Sci USA 2012;109:20089–20094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fjell AM, Grydeland H, Krogsrud SK, et al. . Development and aging of cortical thickness correspond to genetic organization patterns. Proc Natl Acad Sci USA 2015;112:15462–15467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krogsrud SK, Fjell AM, Tamnes CK, et al. . Changes in white matter microstructure in the developing brain—a longitudinal diffusion tensor imaging study of children from 4 to 11 years of age. Neuroimage 2016;124:473–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tamnes CK, Walhovd KB, Grydeland H, et al. . Longitudinal working memory development is related to structural maturation of frontal and parietal cortices. J Cogn Neurosci 2013;25:1611–1623. [DOI] [PubMed] [Google Scholar]
- 17.Storsve AB, Fjell AM, Tamnes CK, et al. . Differential longitudinal changes in cortical thickness, surface area and volume across the adult life span: regions of accelerating and decelerating change. J Neurosci 2014;34:8488–8498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Lange AG, Brathen ACS, Rohani DA, Fjell AM, Walhovd KB. The temporal dynamics of brain plasticity in aging. Cereb Cortex 2018;28:1857–1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Idland AV, Sala-Llonch R, Borza T, et al. . CSF neurofilament light levels predict hippocampal atrophy in cognitively healthy older adults. Neurobiol Aging 2017;49:138–144. [DOI] [PubMed] [Google Scholar]
- 20.Corder EH, Basun H, Lannfelt L, Viitanen M, Winblad B. Attenuation of apolipoprotein E Epsilon4 allele gene dose in late age. Lancet 1996;347:542. [DOI] [PubMed] [Google Scholar]
- 21.Purcell S, Neale B, Todd-Brown K, et al. . PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007;81:559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Auton A, Auton A, Brooks LD, et al. . A global reference for human genetic variation. Nature 2015;526:68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Escott-Price V, Myers AJ, Huentelman M, Hardy J. Polygenic risk score analysis of pathologically confirmed Alzheimer disease. Ann Neurol 2017;82:311–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Escott-Price V, Sims R, Bannister C, et al. . Common polygenic variation enhances risk prediction for Alzheimer's disease. Brain 2015;138:3673–3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Logue MW, Panizzon MS, Elman JA, et al. . Use of an Alzheimer's disease polygenic risk score to identify mild cognitive impairment in adults in their 50s. Mol Psychiatry 2019;24:421–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laws SM, Hone E, Gandy S, Martins RN. Expanding the association between the APOE gene and the risk of Alzheimer's disease: possible roles for APOE promoter polymorphisms and alterations in APOE transcription. J Neurochem 2003;84:1215–1236. [DOI] [PubMed] [Google Scholar]
- 27.Radmanesh F, Devan WJ, Anderson CD, Rosand J, Falcone GJ; Alzheimer's Disease Neuroimaging Initiative. Accuracy of imputation to infer unobserved APOE epsilon alleles in genome-wide genotyping data. Eur J Hum Genet 2014;22:1239–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu C, Kraja AT, Smith JA, et al. . Meta-analysis identifies common and rare variants influencing blood pressure and overlapping with metabolic trait loci. Nat Genet 2016;48:1162–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Patterson N, Price AL, Reich D. Population structure and eigenanalysis. PLoS Genet 2006;2:e190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jovicich J, Czanner S, Greve D, et al. . Reliability in multi-site structural MRI studies: effects of gradient non-linearity correction on phantom and human data. Neuroimage 2006;30:436–443. [DOI] [PubMed] [Google Scholar]
- 31.Fischl B, Salat DH, Busa E, et al. . Whole brain segmentation: automated labeling of neuroanatomical structures in the human brain. Neuron 2002;33:341–355. [DOI] [PubMed] [Google Scholar]
- 32.Knickmeyer RC, Wang J, Zhu H, et al. . Common variants in psychiatric risk genes predict brain structure at birth. Cereb Cortex 2014;24:1230–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cacciaglia R, Molinuevo JL, Falcon C, et al. . APOE-epsilon4 risk variant for Alzheimer's disease modifies the association between cognitive performance and cerebral morphology in healthy middle-aged individuals. Neuroimage Clin 2019;23:101818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Coughlan G, Coutrot A, Khondoker M, Minihane AM, Spiers H, Hornberger M. Toward personalized cognitive diagnostics of at-genetic-risk Alzheimer's disease. Proc Natl Acad Sci USA 2019;116:9285–9292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lindenberger U, Nagel IE, Chicherio C, Li SC, Heekeren HR, Bäckman L. Age-related decline in brain resources modulates genetic effects on cognitive functioning. Front Neurosci 2008;2:234–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nagel IE, Chicherio C, Li SC, et al. . Human aging magnifies genetic effects on executive functioning and working memory. Front Hum Neurosci 2008;2:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Papenberg G, Lindenberger U, Backman L. Aging-related magnification of genetic effects on cognitive and brain integrity. Trends Cogn Sci 2015;19:506–514. [DOI] [PubMed] [Google Scholar]
- 38.Katzman R, Terry R, DeTeresa R, et al. . Clinical, pathological, and neurochemical changes in dementia: a subgroup with preserved mental status and numerous neocortical plaques. Ann Neurol 1988;23:138–144. [DOI] [PubMed] [Google Scholar]
- 39.Stern Y. Cognitive reserve in ageing and Alzheimer's disease. Lancet Neurol 2012;11:1006–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Livingston G, Sommerlad A, Orgeta V, et al. . Dementia prevention, intervention, and care. Lancet 2017;390:2673–2734. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
There are data access restrictions in accordance with ethical guidelines and national data protection legislation. The data will be made available pending reasonable request, appropriate institutional data protection security measures, and ethical approval.