Fig. 3. Variability in CSF PGRN levels explained by genetic variants.
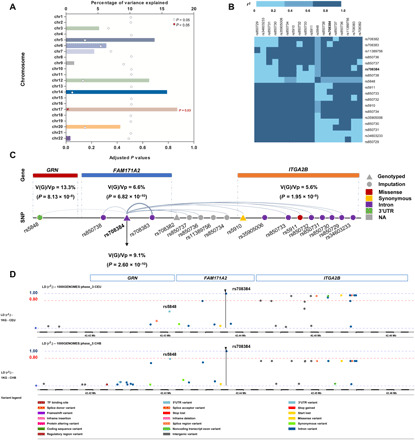
(A) Chromosome 17 explained approximately 17.4% of the variability in the CSF levels of PGRN. (B) Most of the significant loci were in LD with rs708384. (C) SNPs in the GRN region explained the most but not all of the variability in CSF PGRN. Analysis of FAM171A2 and ITGA2B region showed that these two regions explained 9.1 and 5.6% of the variability in CSF levels of PGRN, respectively. rs708384 was shown to explain 9.1% of the variability. (D) rs5848 was only in low-to-moderate LD with rs708384 in CEU population (r2 = 0.6), and no loci were in LD with rs5848. Nonetheless, rs5848 was in high LD with rs708384 in CHB population (r2 ≈ 0.8).
