Extended Data Figure 7. Context specific requirement for GPC3 in glioma tumorigenesis.
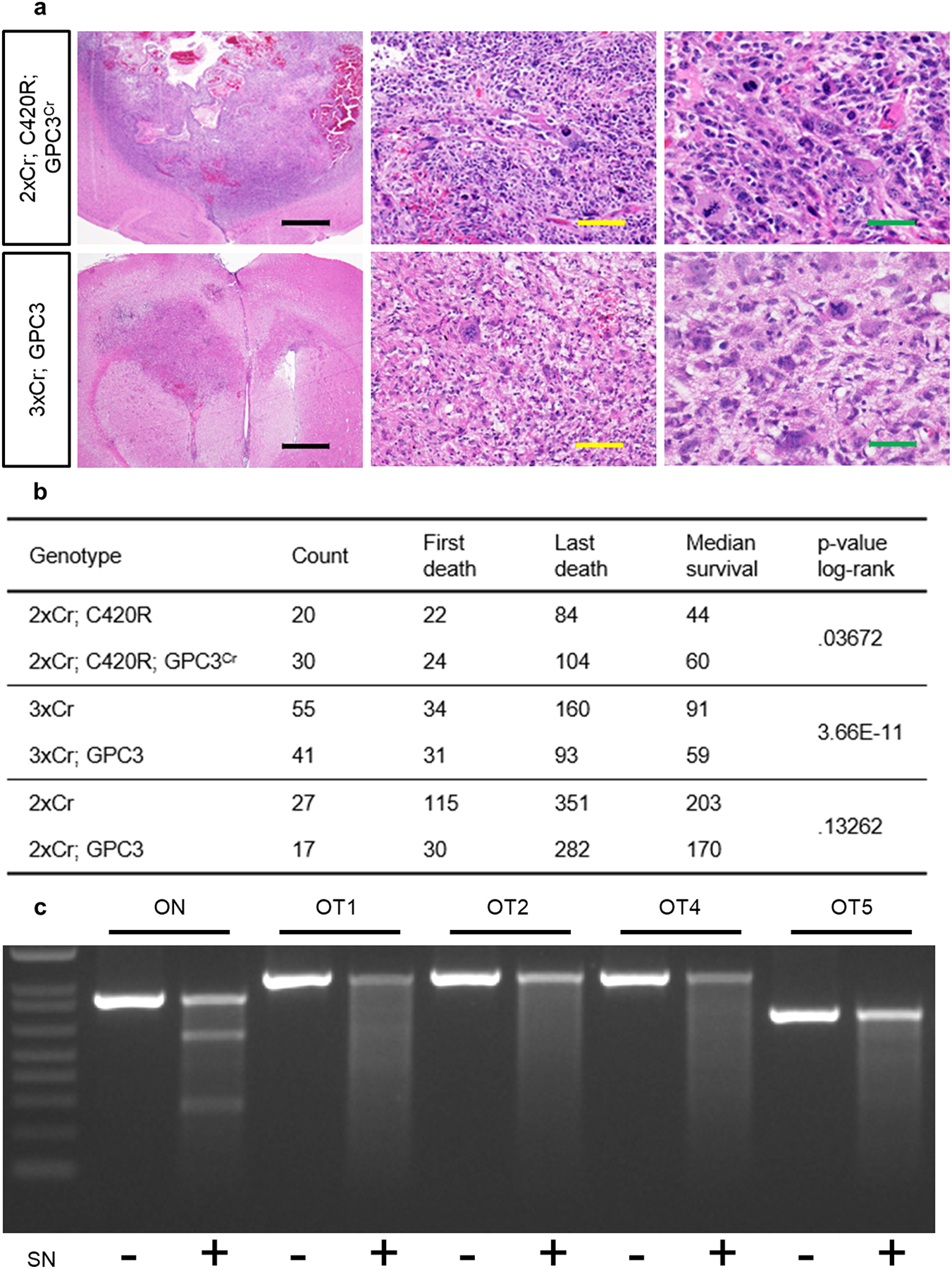
a) H&E staining of 2xCr; C420R; GPC3Cr (top) and 3xCr; GPC3 (bottom) tumor brains histologically graded as high grade glioma, either WHO grade III anaplastic astrocytoma or Grade IV GBM. Black scale bar = 1mm; yellow scale bar = 100μm; green scale bar = 50μm. Representative images of each variant driven tumor are from N = 6 brains. b) Survival statistics for in vivo modeling GPC3 loss and gain in various tumor models. p-values were calculated using the log rank test. c) SURVEYOR assay analysis on genomic DNA from 2xCr; C420R; GPC3Cr tissue. Representative DNA gel electrophoresis of PCR products after SURVEYOR enzyme treatment. After the ladder (1KB Plus, ThermoFisher, 10787018) ranging from 100bp – 1500bp, lanes contain on-target (ON) and top 5 off-target (OT) sites with and without nuclease treatment. OT3 rests in an AT rich region and was not amplifiable. This experiment was independently repeated 3 times with similar results. d) Table lists the on and off target sequences of the GPC3 CRISPR gRNA sequence. Off target sequences denote mismatches (lowercase) or bulges (−). In addition to the genomic location of sites, listed are primer sequences used to amplify target regions. Off target site 3 was not amplifiable because it was an AT-rich region, not amendable to specific primers.
