Figure 3. Example of a spatiotemporal optogenetics experiment with subcellular resolution.
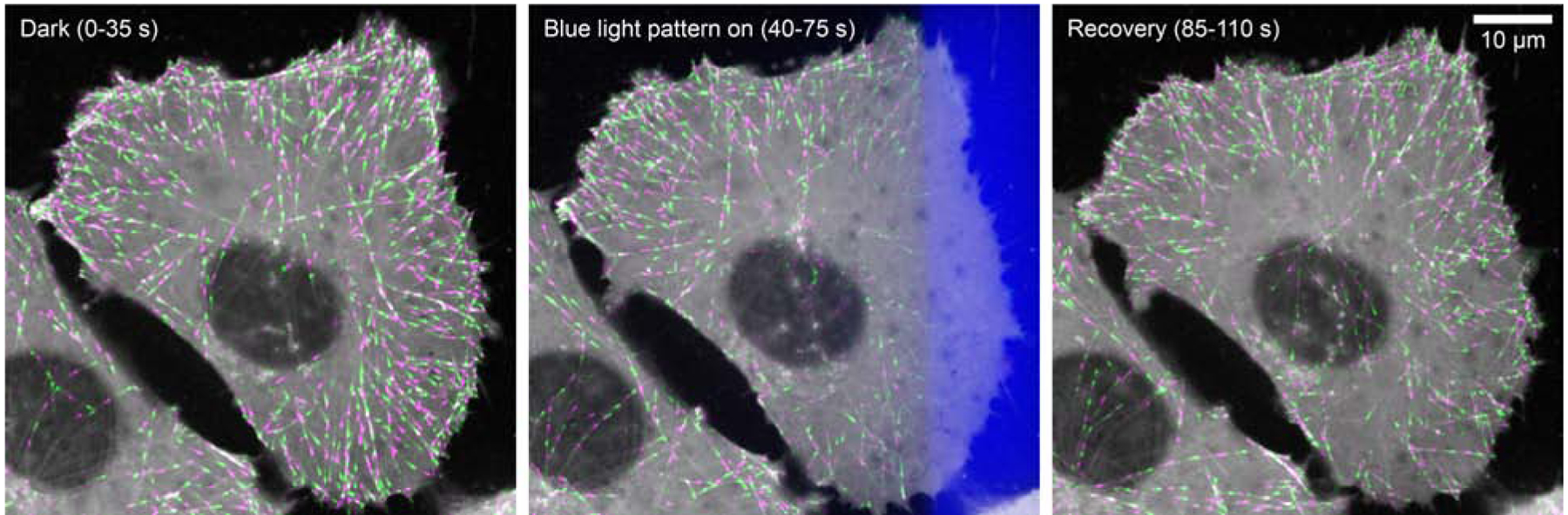
Shown is a human tissue culture cell in which a photosensitive LOV2/Zdk1 protein-protein interaction module was inserted into the endogenous EB1 gene by CRISPR/Cas9 genome editing, which replaces the wild-type EB1 protein with the light sensitive π-EB1 variant in one step. The images show an overlay of the mCherry-tagged π-EB1 C-terminal half on growing microtubule ends from short time-lapse sequences (8 images every 5 seconds) in alternating green and magenta before, during and after localized 470 nm blue light exposure. The light-exposed region is indicated by the blue overlay in the middle panel. Note the gradient of π-EB1 dissociation near the edge of the light-exposed region that results from diffusion of photoactivated molecules. Sharper boundaries can be achieved with LOV2 domain variants with faster dark recovery rates [45].
